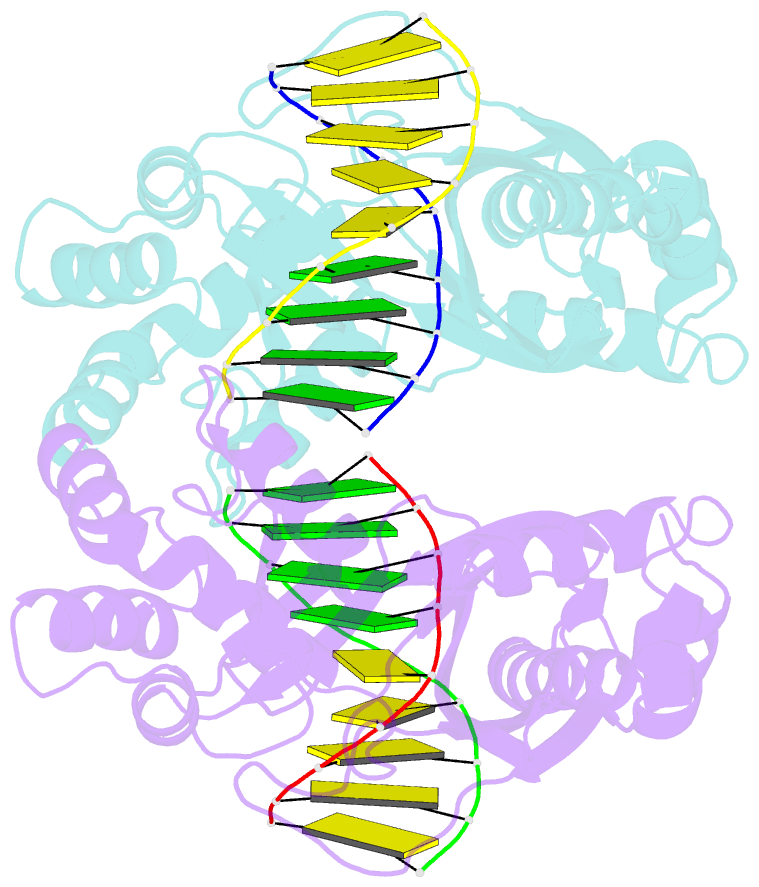
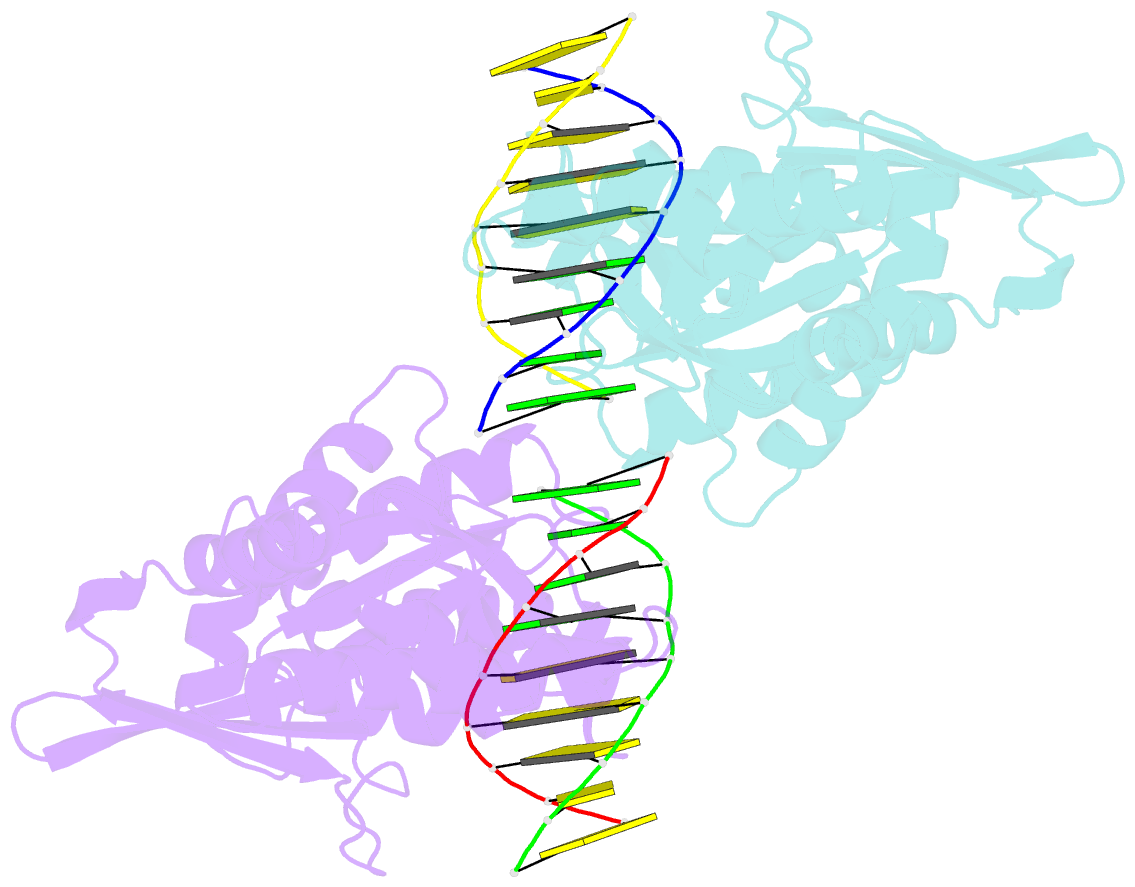
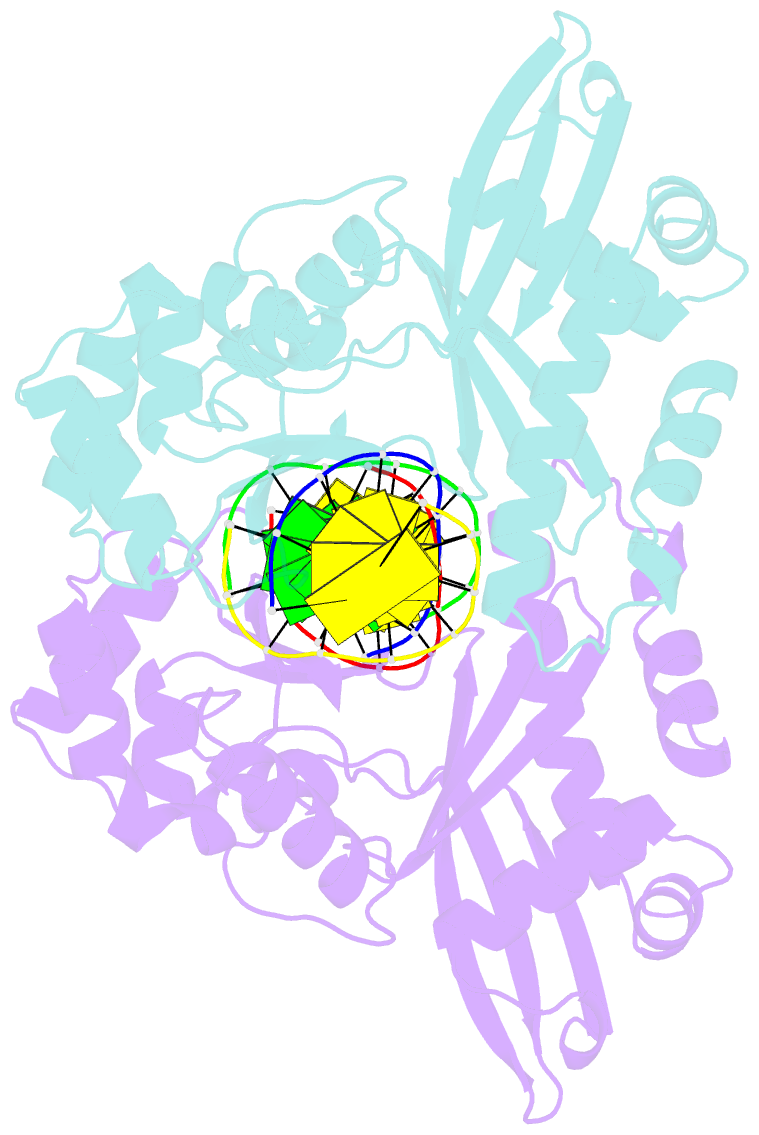
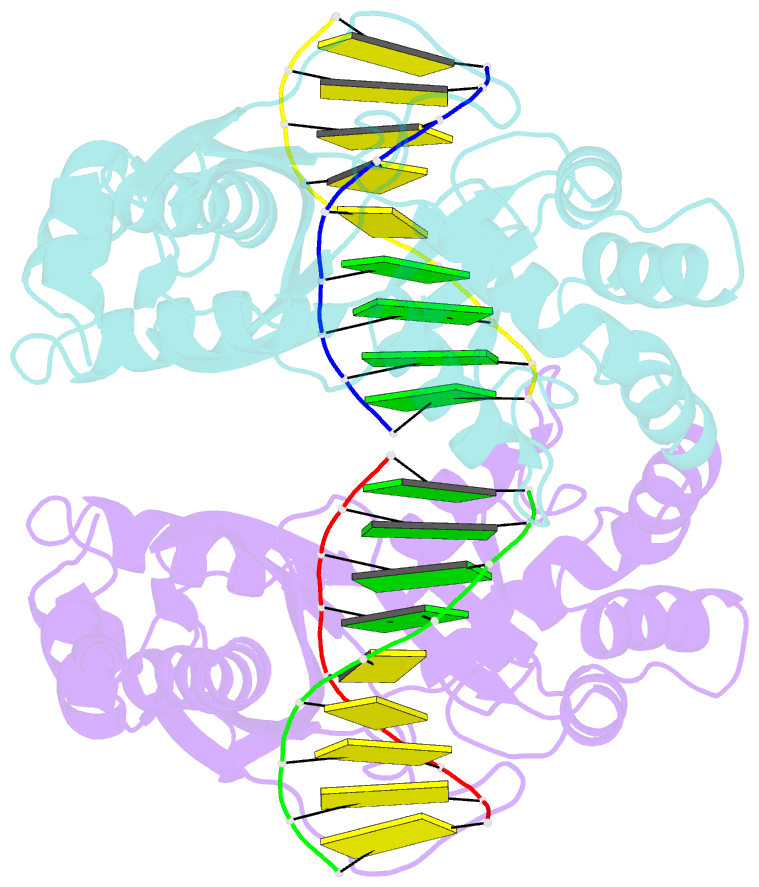
Summary information and primary citation
- PDB-id
- 1sa3; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- hydrolase-DNA
- Method
- X-ray (1.95 Å)
- Summary
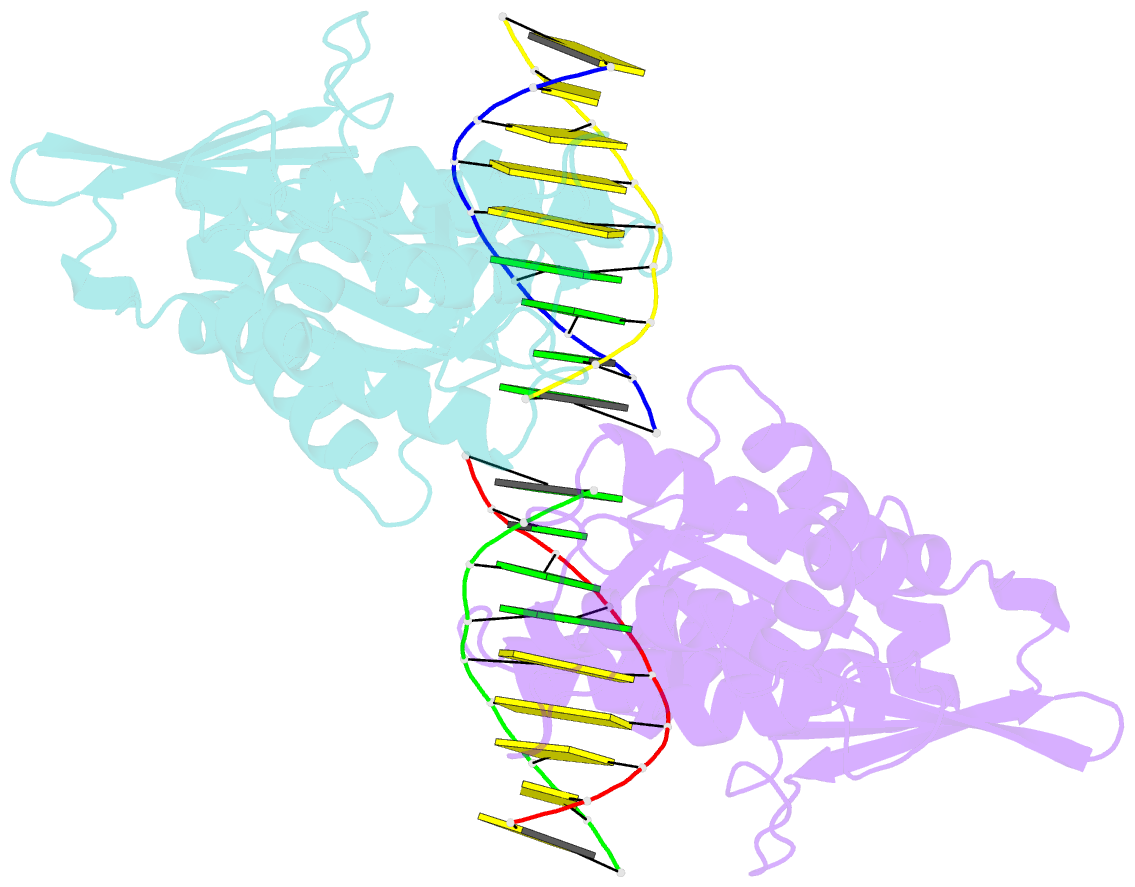
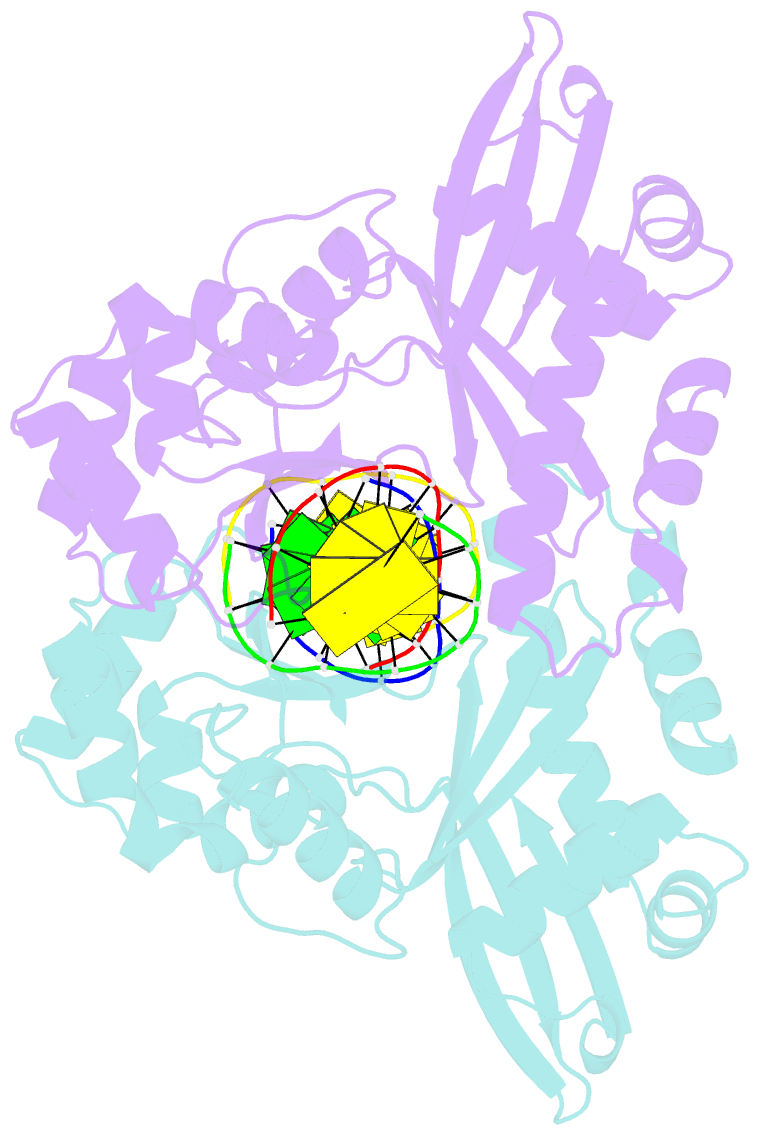
- An asymmetric complex of restriction endonuclease mspi on its palindromic DNA recognition site
- Reference
- Xu QS, Kucera RB, Roberts RJ, Guo HC (2004): "An Asymmetric Complex of Restriction Endonuclease MspI on Its Palindromic DNA Recognition Site." STRUCTURE, 12, 1741-1747. doi: 10.1016/j.str.2004.07.014.
- Abstract
- Most well-known restriction endonucleases recognize palindromic DNA sequences and are classified as Type IIP. Due to the recognition and cleavage symmetry, Type IIP enzymes are usually found to act as homodimers in forming 2-fold symmetric enzyme-DNA complexes. Here we report an asymmetric complex of the Type IIP restriction enzyme MspI in complex with its cognate recognition sequence. Unlike any other Type IIP enzyme reported to date, an MspI monomer and not a dimer binds to a palindromic DNA sequence. The enzyme makes specific contacts with all 4 base pairs in the recognition sequence, by six direct and five water-mediated hydrogen bonds and numerous van der Waal contacts. This MspI-DNA structure represents the first example of asymmetric recognition of a palindromic DNA sequence by two different structural motifs in one polypeptide. A few possible pathways are discussed for MspI to cut both strands of DNA, either as a monomer or dimer.