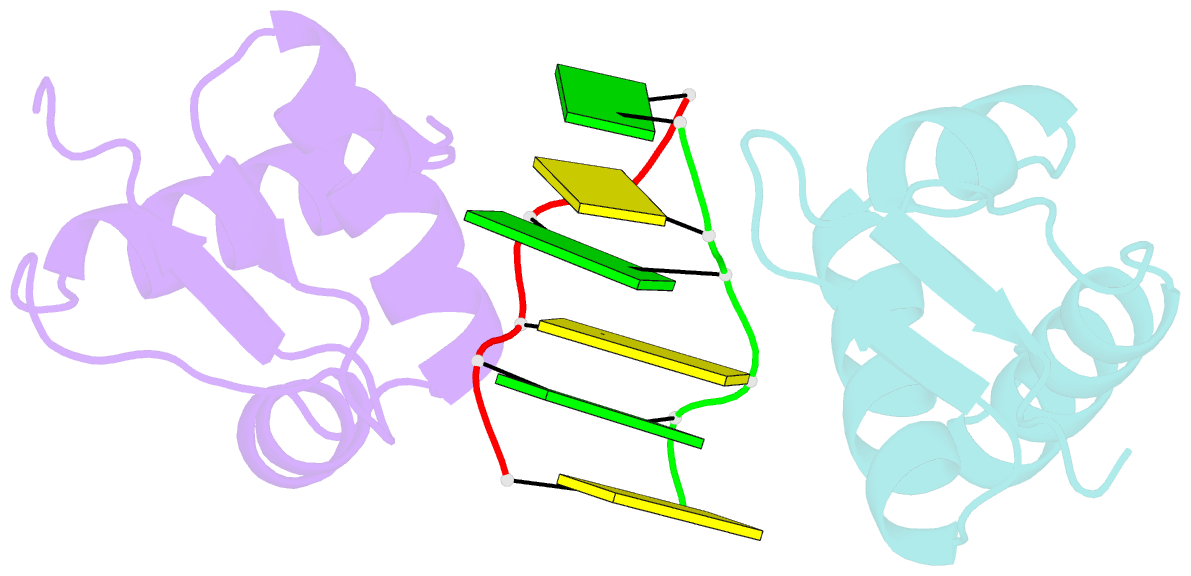
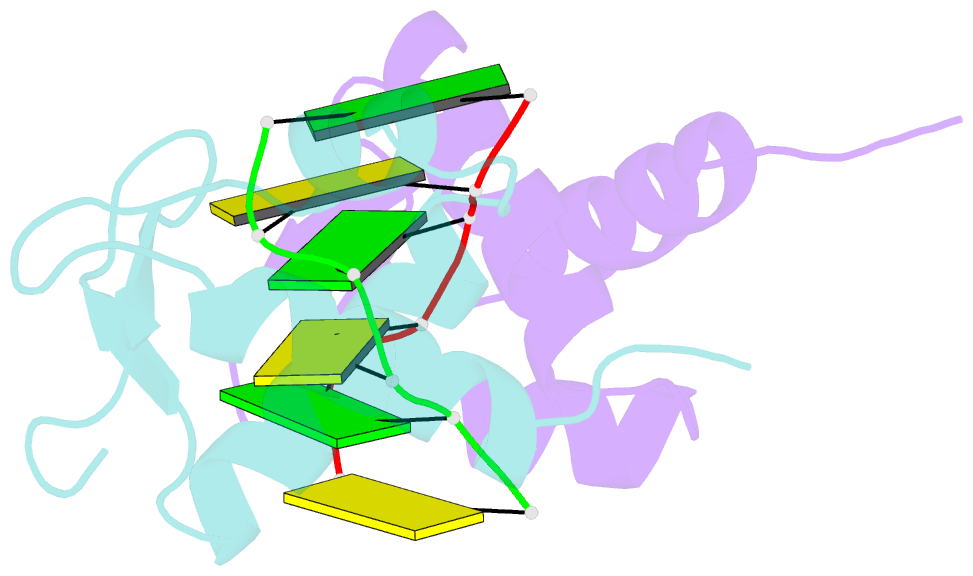
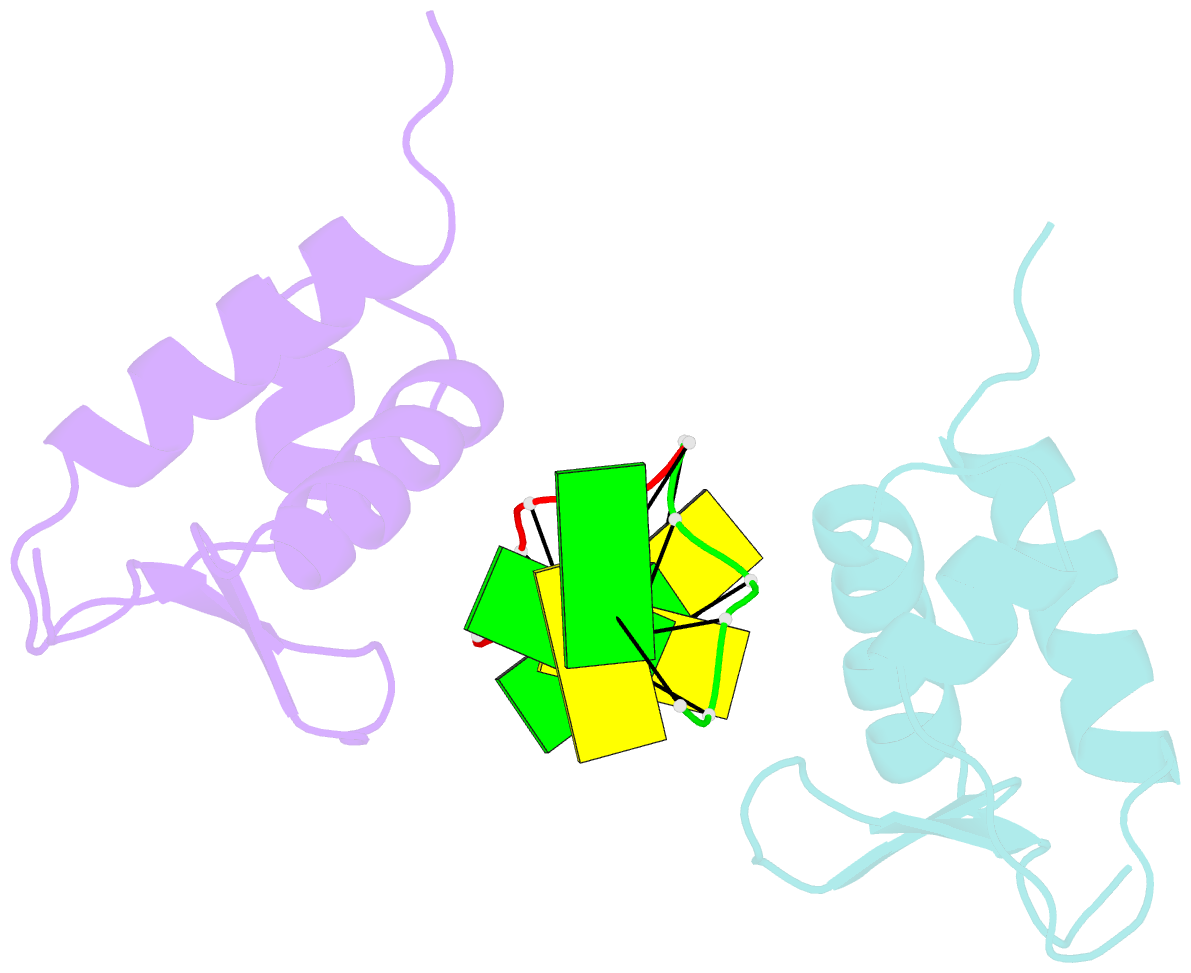
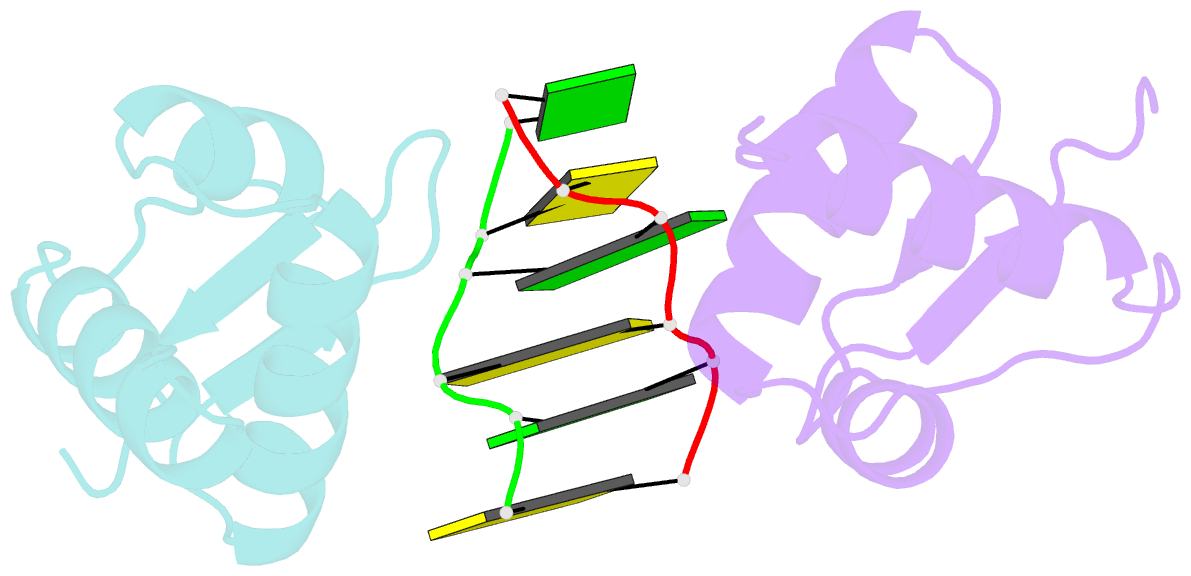
Summary information and primary citation
- PDB-id
- 1sfu; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- DNA binding protein-DNA
- Method
- X-ray (2.0 Å)
- Summary
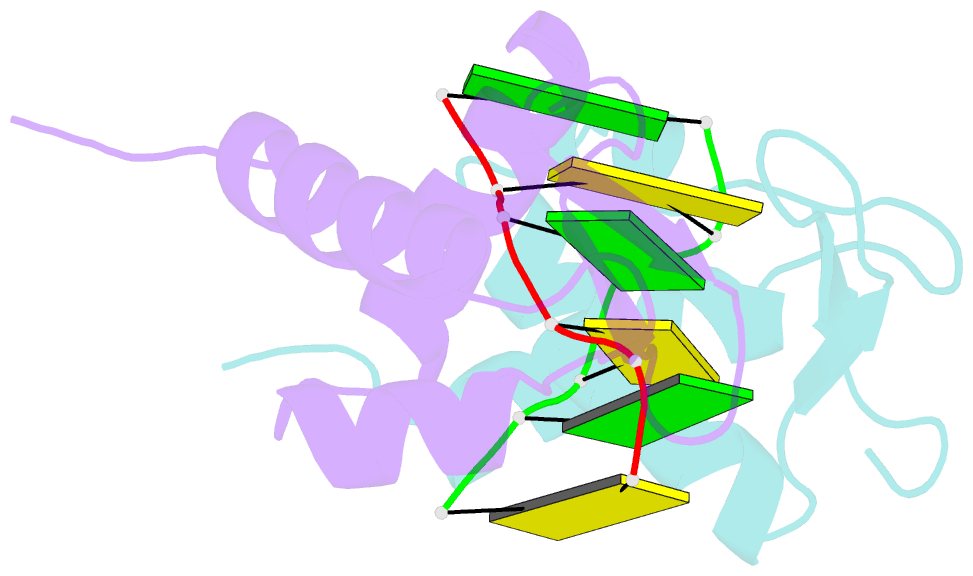
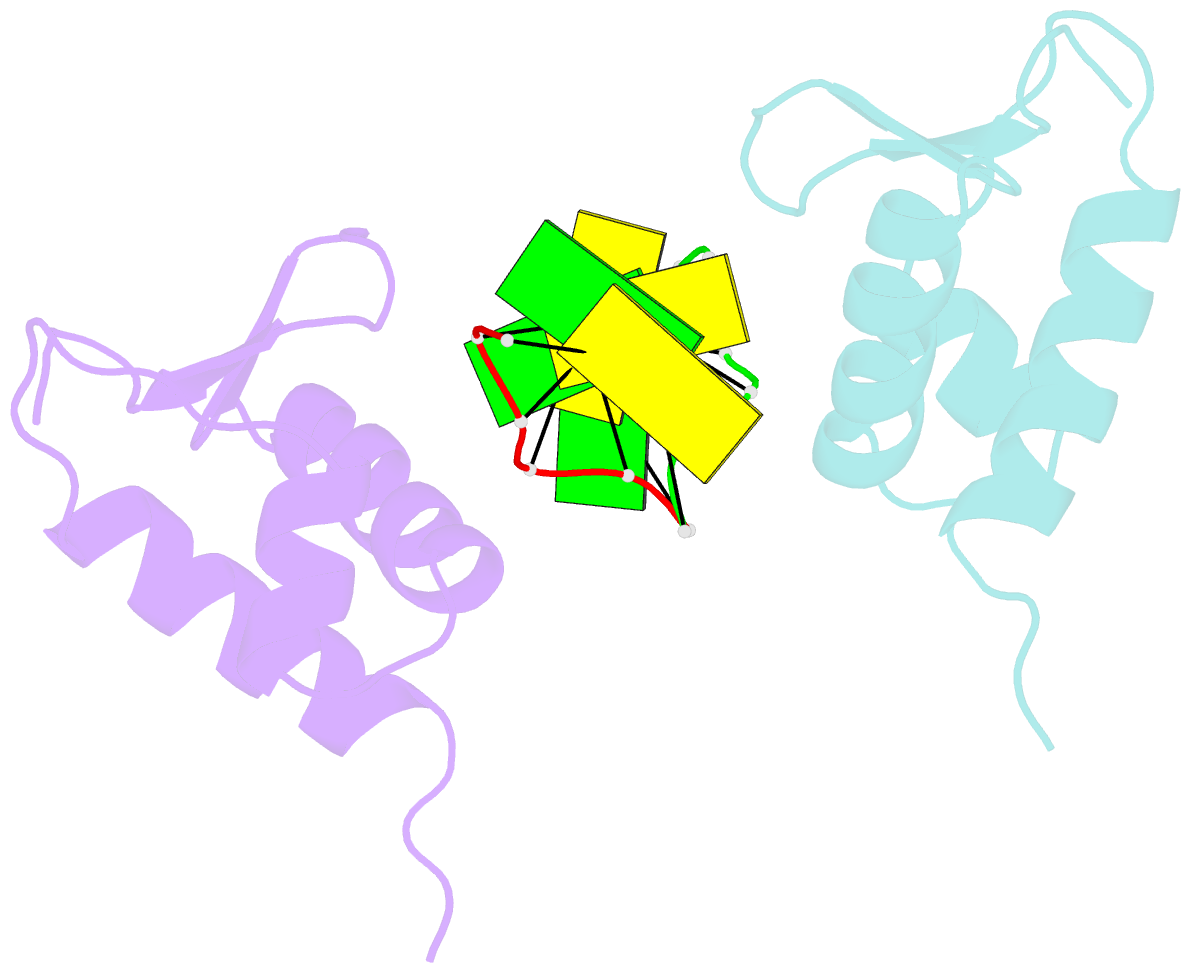
- Crystal structure of the viral zalpha domain bound to left-handed z-DNA
- Reference
- Ha SC, Lokanath NK, Van Quyen D, Wu CA, Lowenhaupt K, Rich A, Kim YG, Kim KK (2004): "A poxvirus protein forms a complex with left-handed Z-DNA: crystal structure of a Yatapoxvirus Zalpha bound to DNA." Proc.Natl.Acad.Sci.USA, 101, 14367-14372. doi: 10.1073/pnas.0405586101.
- Abstract
- A conserved feature of poxviruses is a protein, well characterized as E3L in vaccinia virus, that confers IFN resistance on the virus. This protein comprises two domains, an N-terminal Z-DNA-binding protein domain (Zalpha) and a C-terminal double-stranded RNA-binding domain. Both are required for pathogenicity of vaccinia virus in mice infected by intracranial injection. Here, we describe the crystal structure of the Zalpha domain from the E3L-like protein of Yaba-like disease virus, a Yatapoxvirus, in a complex with Z-DNA, solved at a 2.0-A resolution. The DNA contacting surface of Yaba-like disease virus Zalpha(E3L) closely resembles that of other structurally defined members of the Zalpha family, although some variability exists in the beta-hairpin region. In contrast to the Z-DNA-contacting surface, the nonbinding surface of members of the Zalpha family are unrelated; this surface may effect protein-specific interactions. The presence of the conserved and tailored Z-DNA-binding surface, which interacts specifically with the zigzag backbone and syn base diagnostic of the Z-form, reinforces the importance to poxvirus infection of the ability of this protein to recognize the Z-conformation.