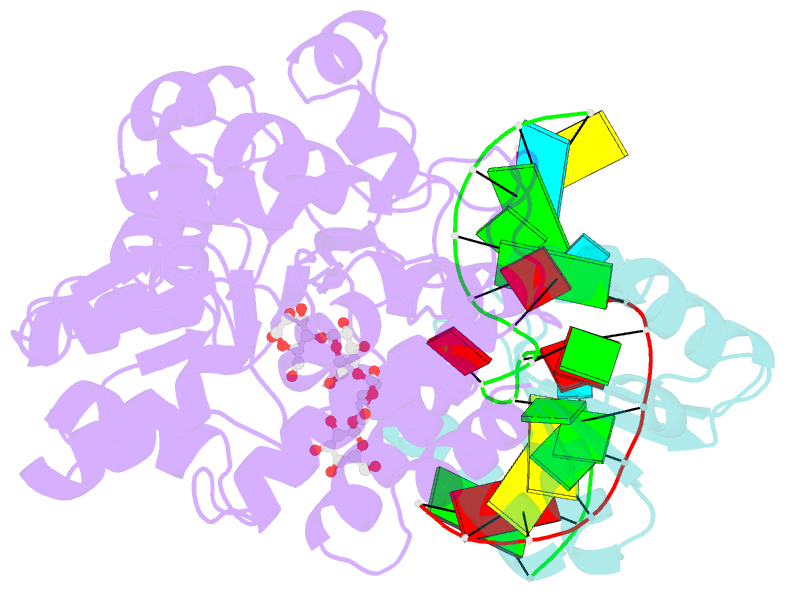
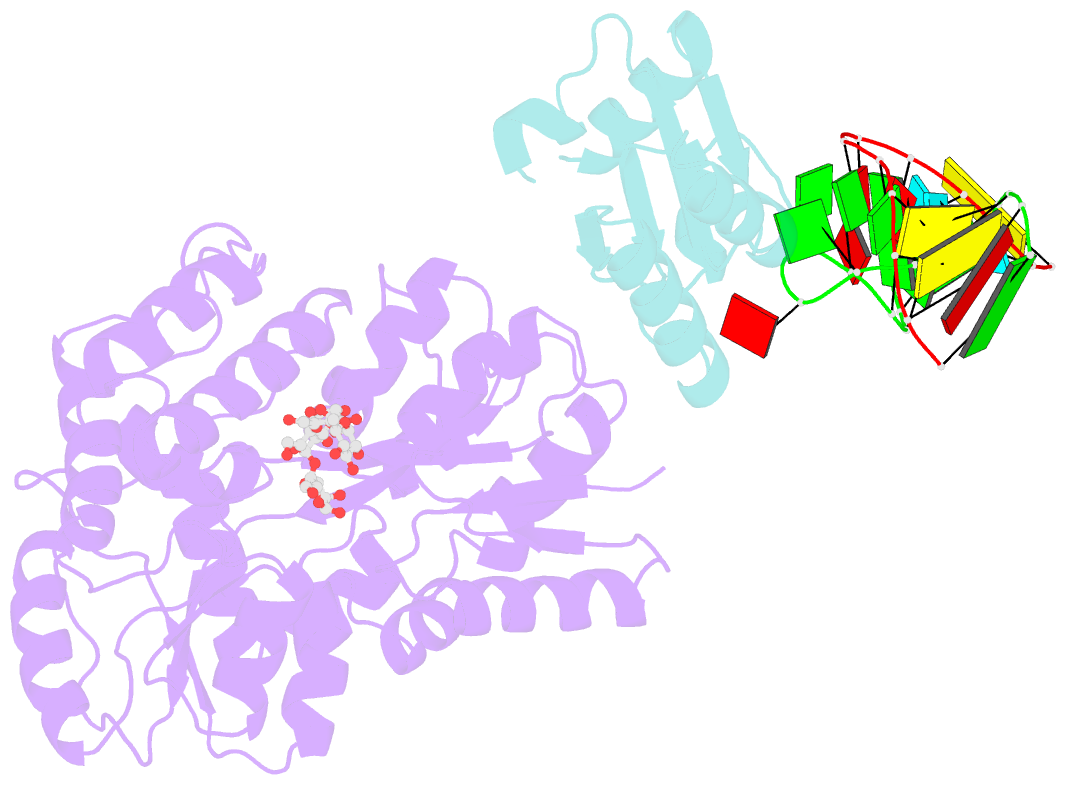
Summary information and primary citation
- PDB-id
- 1t0k; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- ribosome
- Method
- X-ray (3.24 Å)
- Summary
- Joint x-ray and NMR refinement of yeast l30e-mrna complex
- Reference
- Chao JA, Williamson JR (2004): "Joint X-Ray and NMR Refinement of the Yeast L30e-mRNA Complex." Structure, 12, 1165-1176. doi: 10.1016/j.str.2004.04.023.
- Abstract
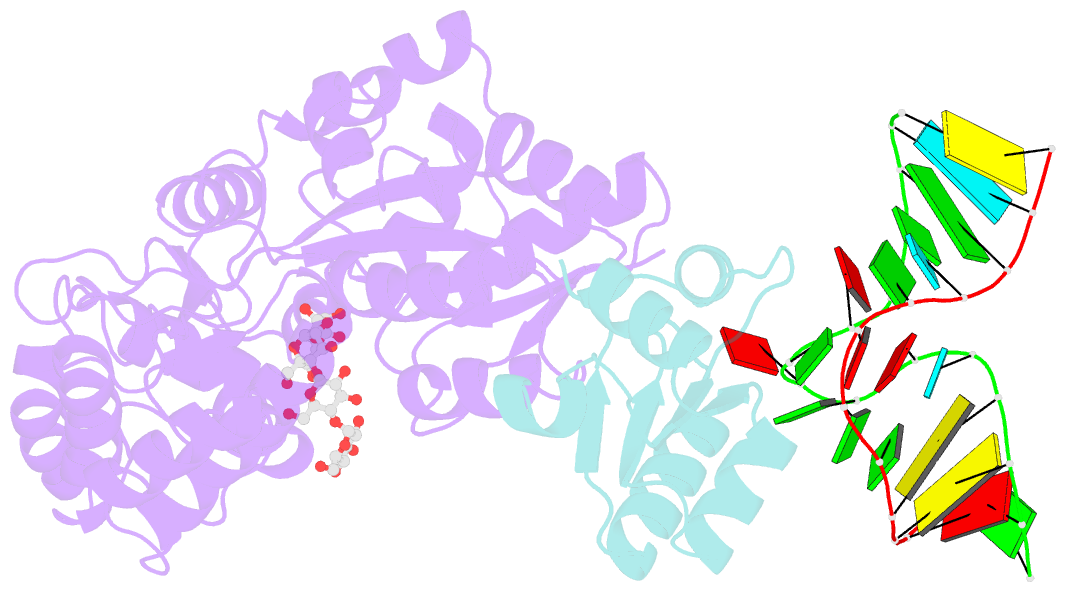
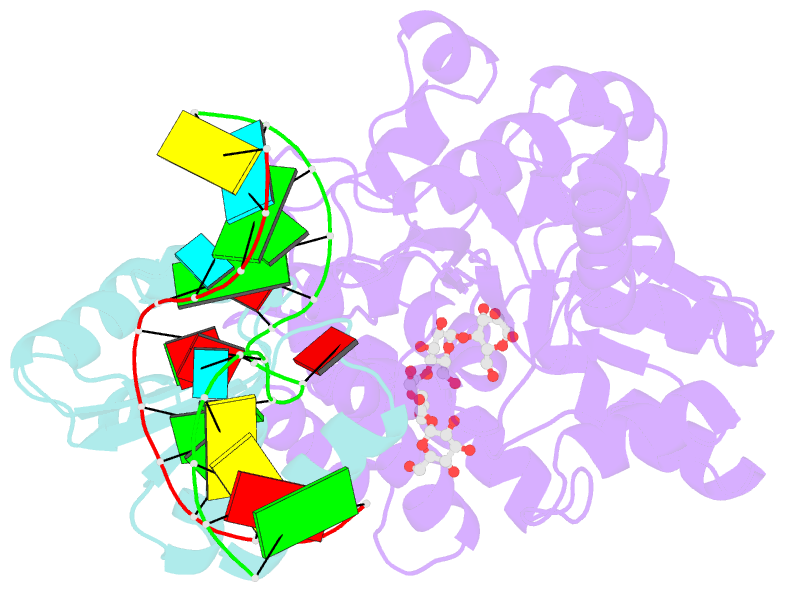
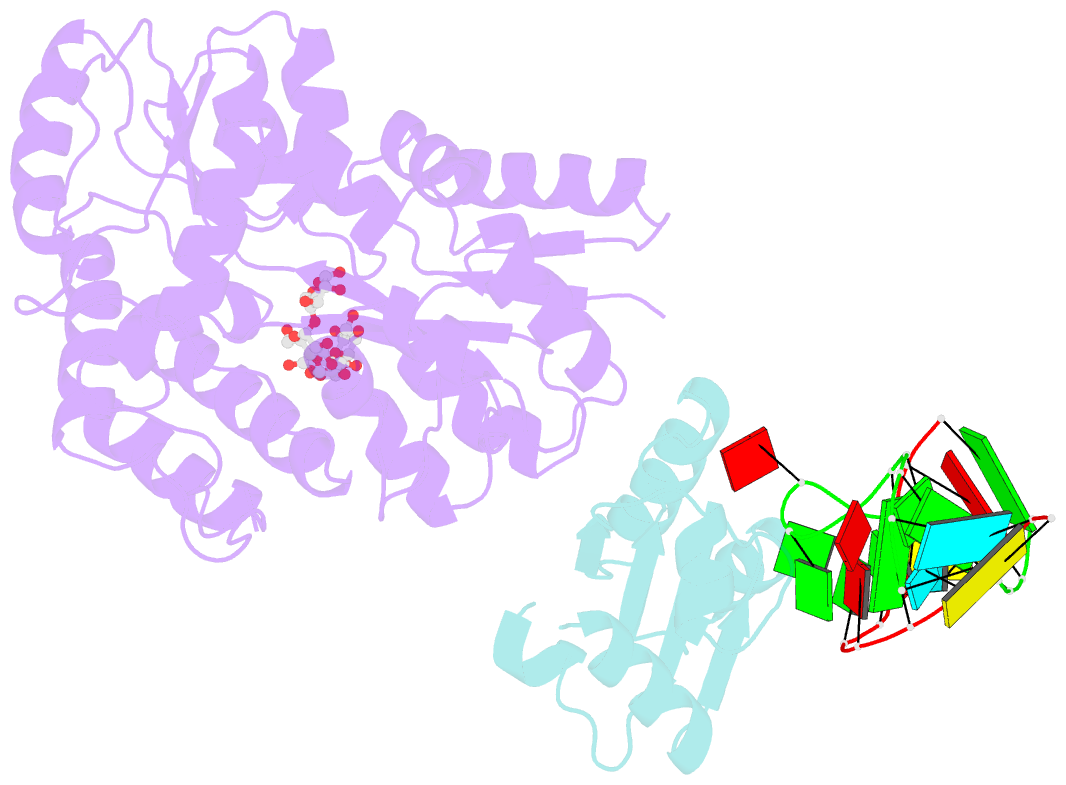
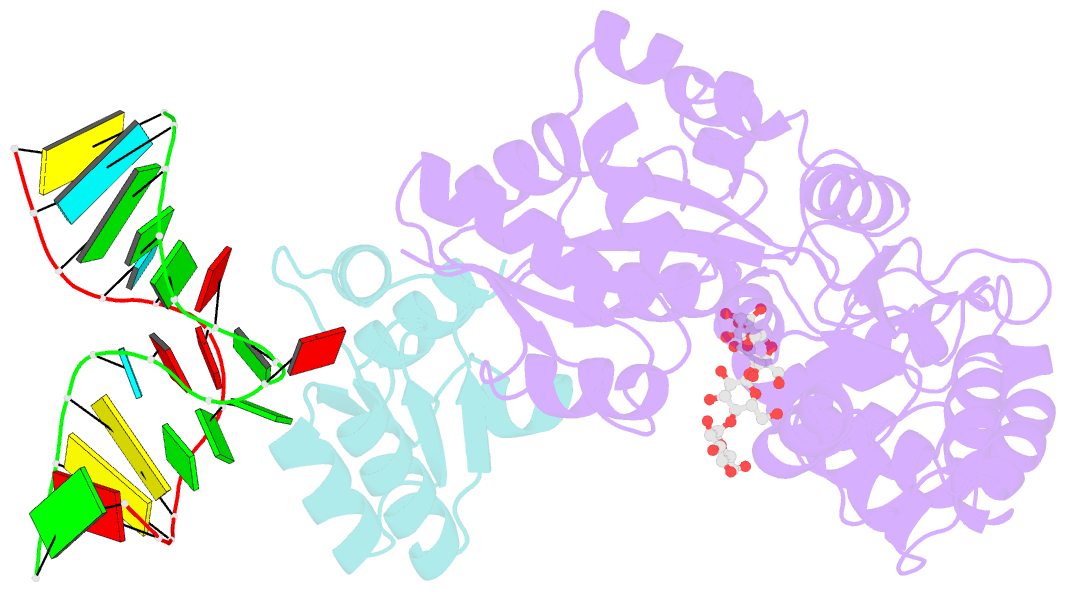
- L30e, a Saccharomyces cervisiae ribosomal protein, regulates its own expression by binding to a purine-rich asymmetric internal loop located in both its pre-mRNA and mature mRNA. A crystal structure of an MBP-L30e fusion protein in complex with an RNA containing the pre-mRNA regulatory site was solved at 3.24 A. Interestingly, the structure of the RNA differed from that observed in a previously determined NMR structure of the complex. Analysis of the NMR data led to the identification of a single imino proton resonance in the internal loop that had been incorrectly assigned and was principally responsible for the erroneous RNA structure. A structure refinement was performed using both the X-ray diffraction data and the NMR-derived distance and angle restraints. The joint NMR and X-ray refinement resulted in improved stereochemistry and lower crystallographic R factors. The RNA internal loop of the MBP-L30e-mRNA complex adopts the canonical K-turn fold.