Summary information and primary citation
- PDB-id
- 1t7p; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transferase-DNA
- Method
- X-ray (2.2 Å)
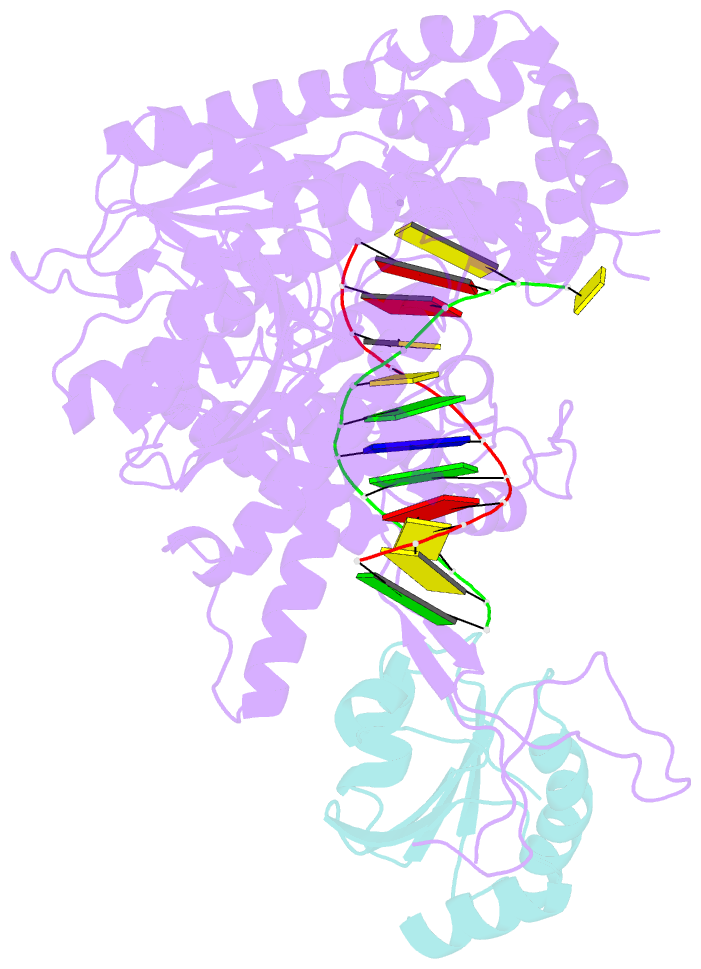
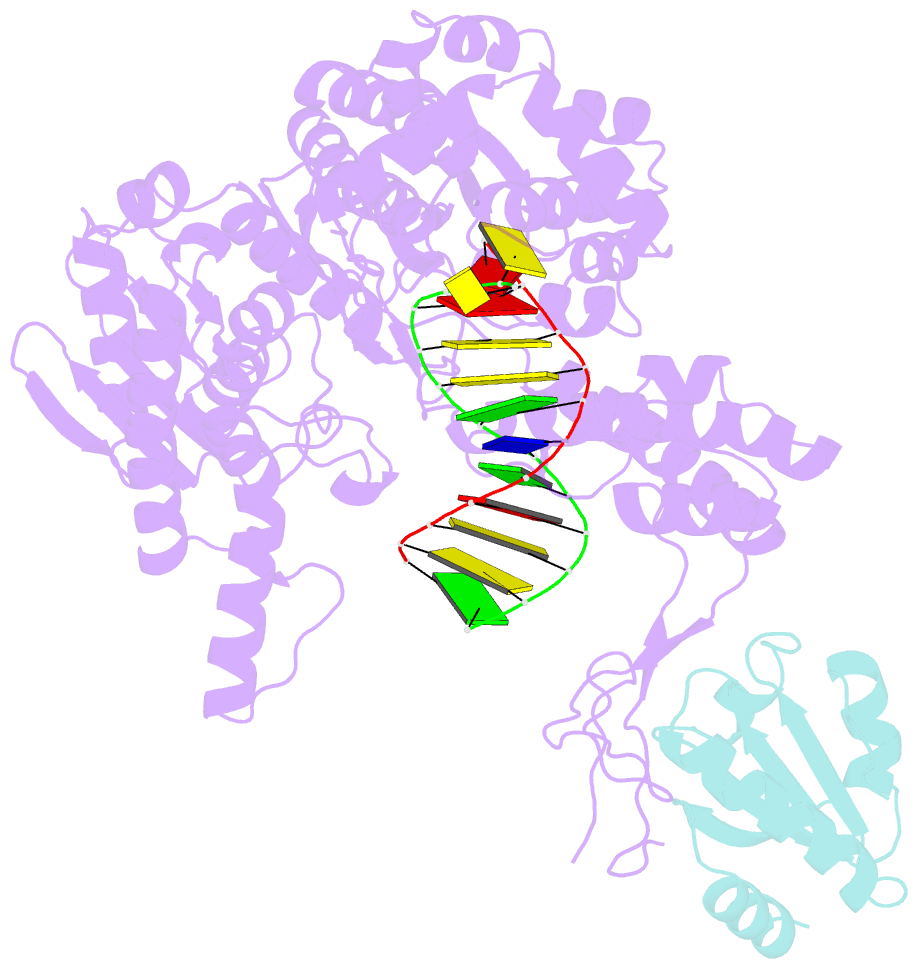
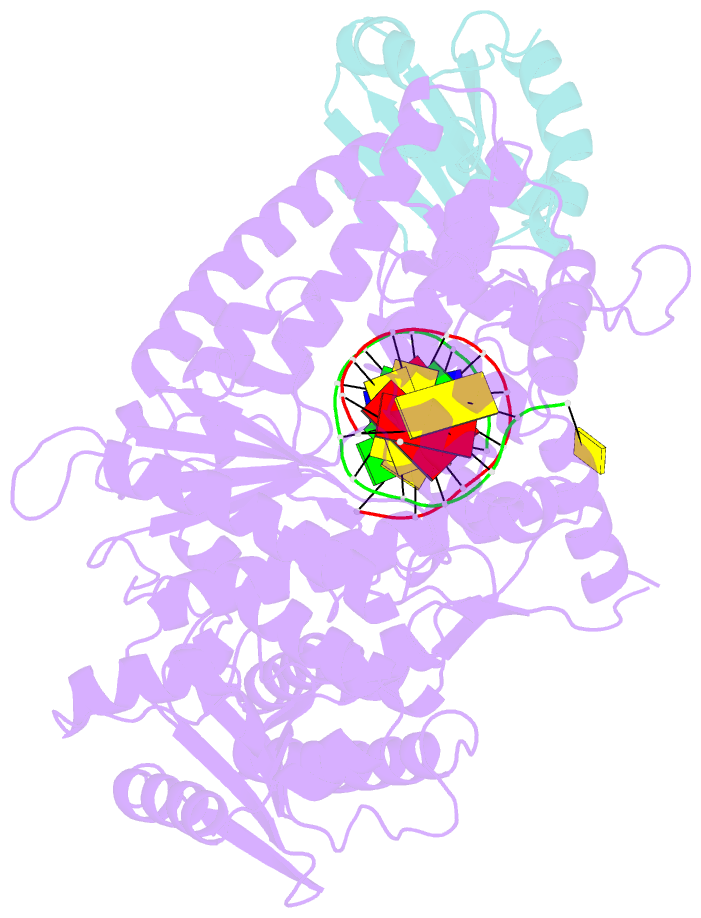
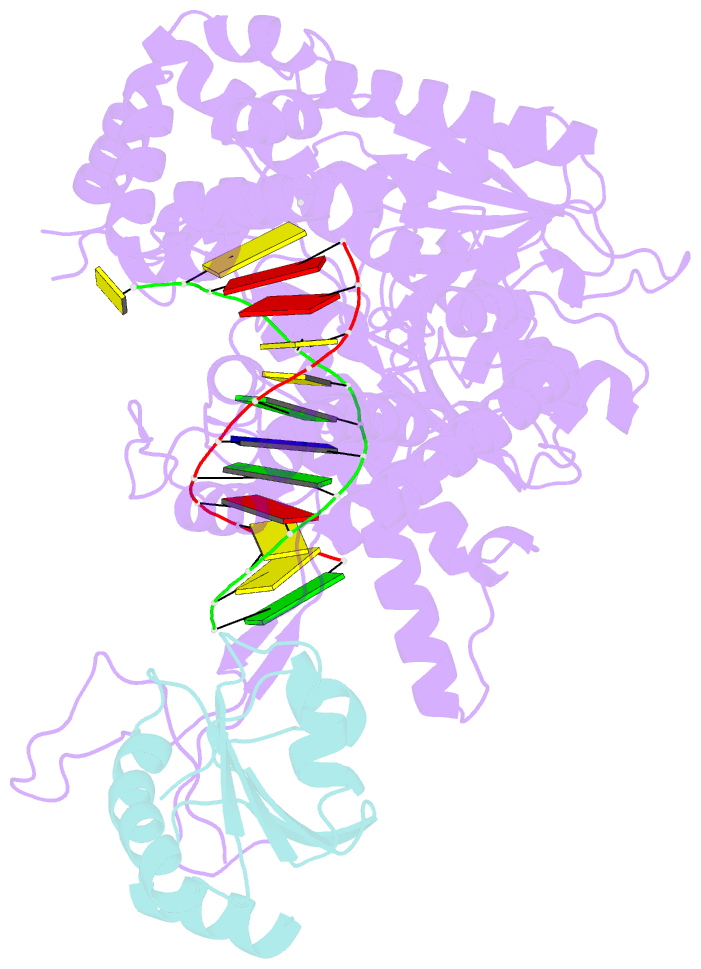
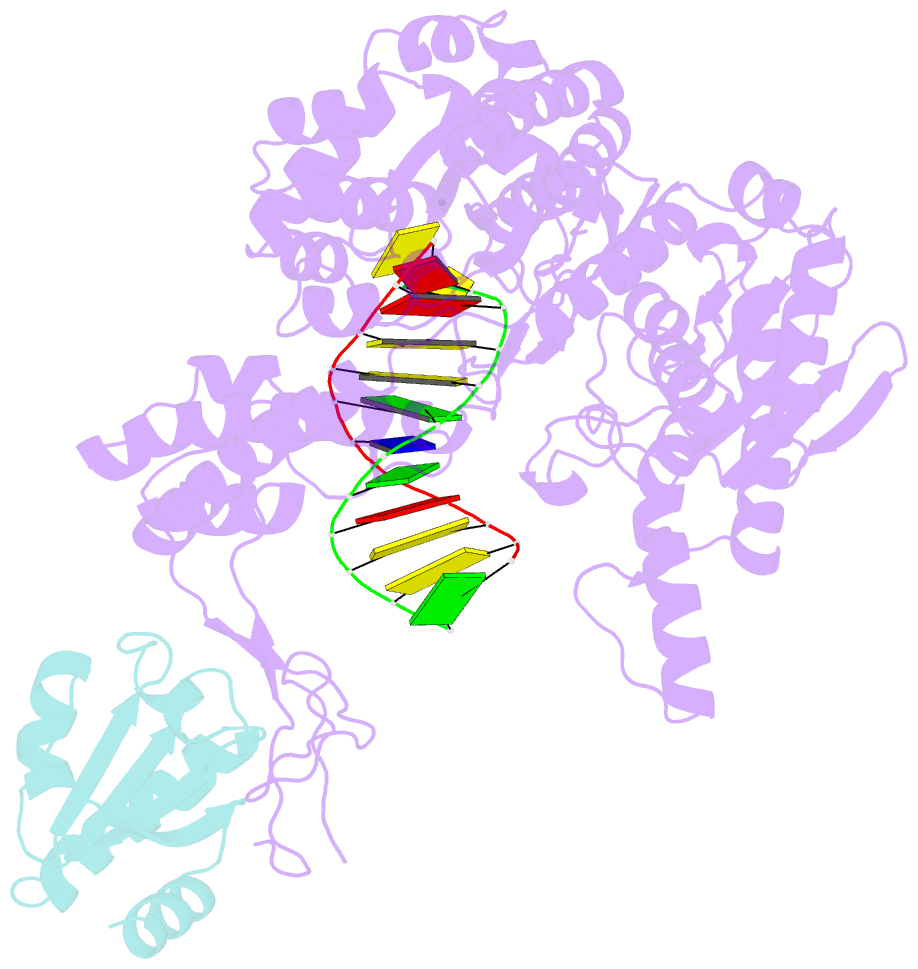
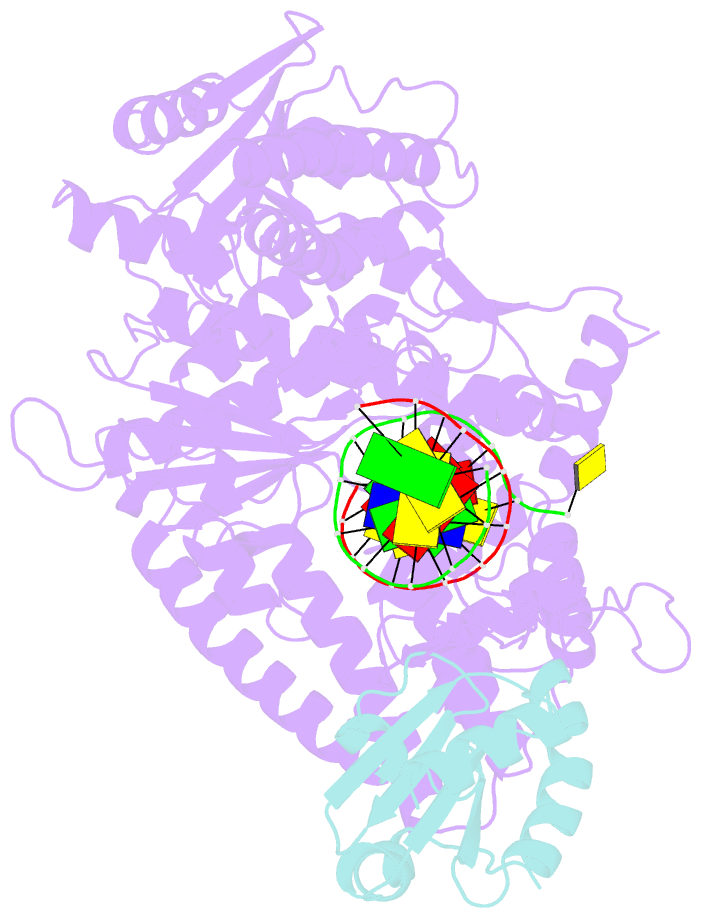
- Summary
- T7 DNA polymerase complexed to DNA primer-template,a nucleoside triphosphate, and its processivity factor thioredoxin
- Reference
- Doublie S, Tabor S, Long AM, Richardson CC, Ellenberger T (1998): "Crystal structure of a bacteriophage T7 DNA replication complex at 2.2 A resolution." Nature, 391, 251-258. doi: 10.1038/34593.
- Abstract
- DNA polymerases change their specificity for nucleotide substrates with each catalytic cycle, while achieving error frequencies in the range of 10(-5) to 10(-6). Here we present a 2.2 A crystal structure of the replicative DNA polymerase from bacteriophage T7 complexed with a primer-template and a nucleoside triphosphate in the polymerase active site. The structure illustrates how nucleotides are selected in a template-directed manner, and provides a structural basis for a metal-assisted mechanism of phosphoryl transfer by a large group of related polymerases.