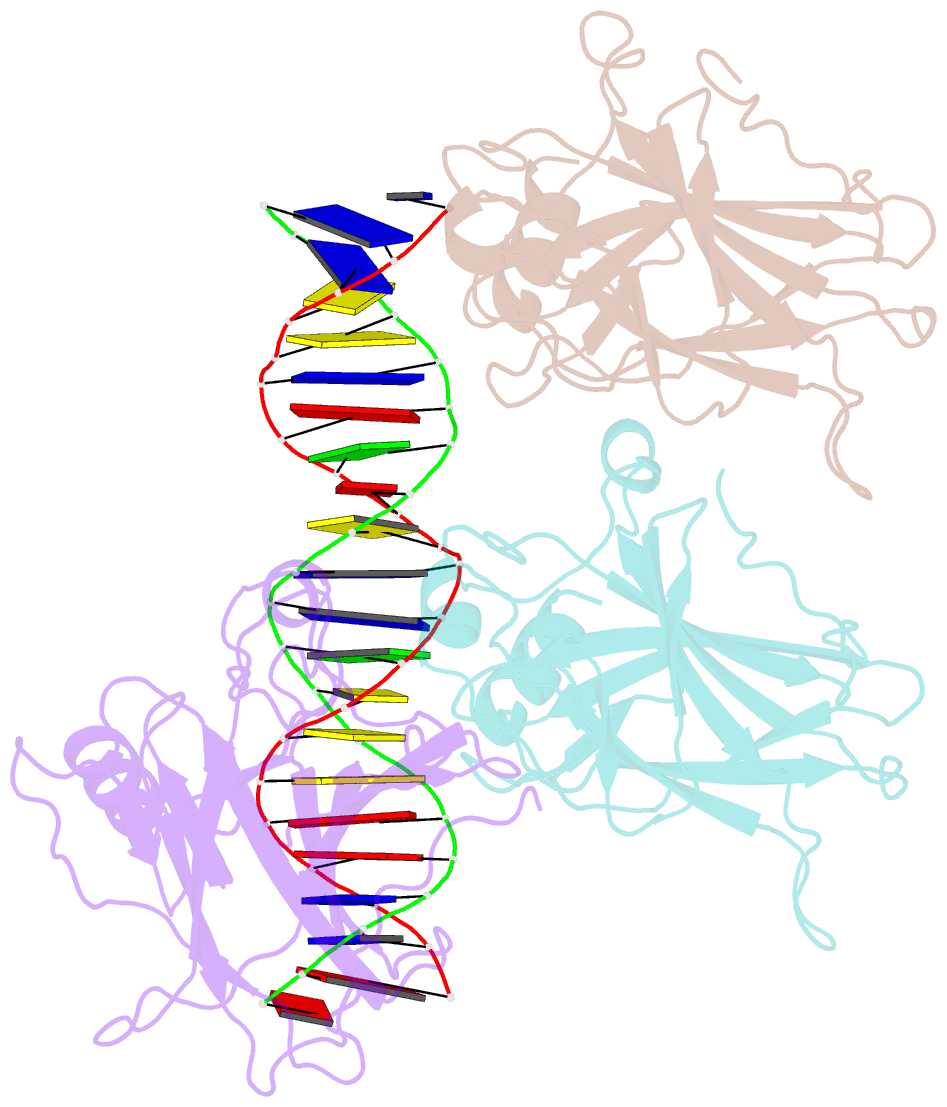
Summary information and primary citation
- PDB-id
- 1tup; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- antitumor protein-DNA
- Method
- X-ray (2.2 Å)
- Summary
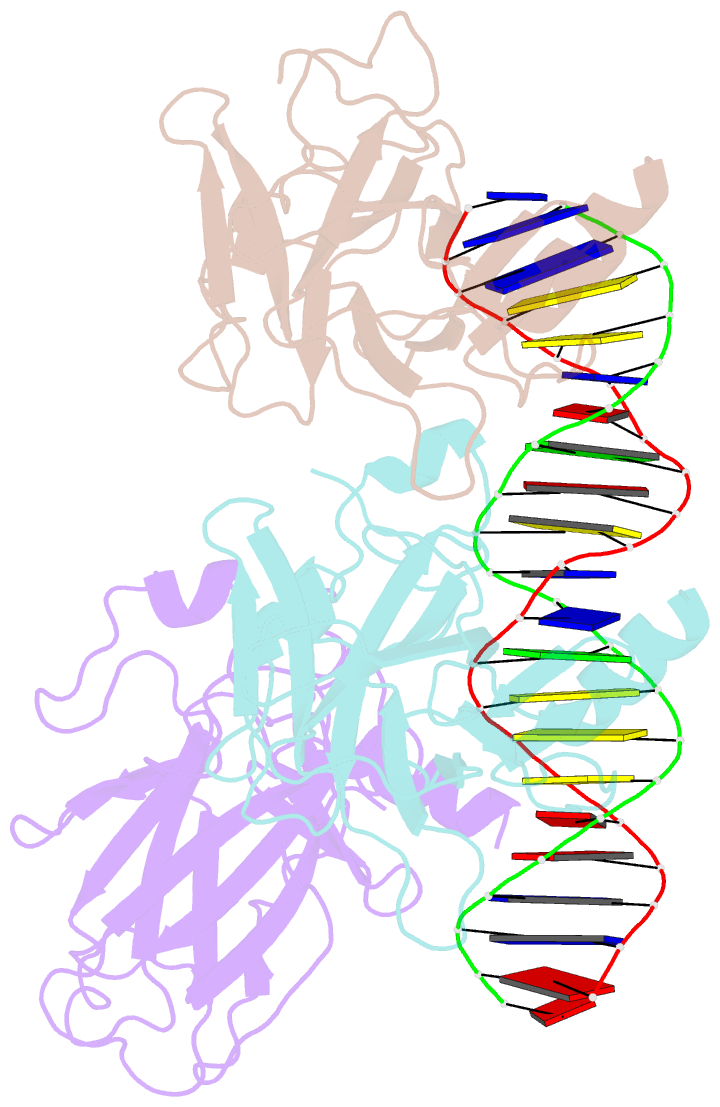
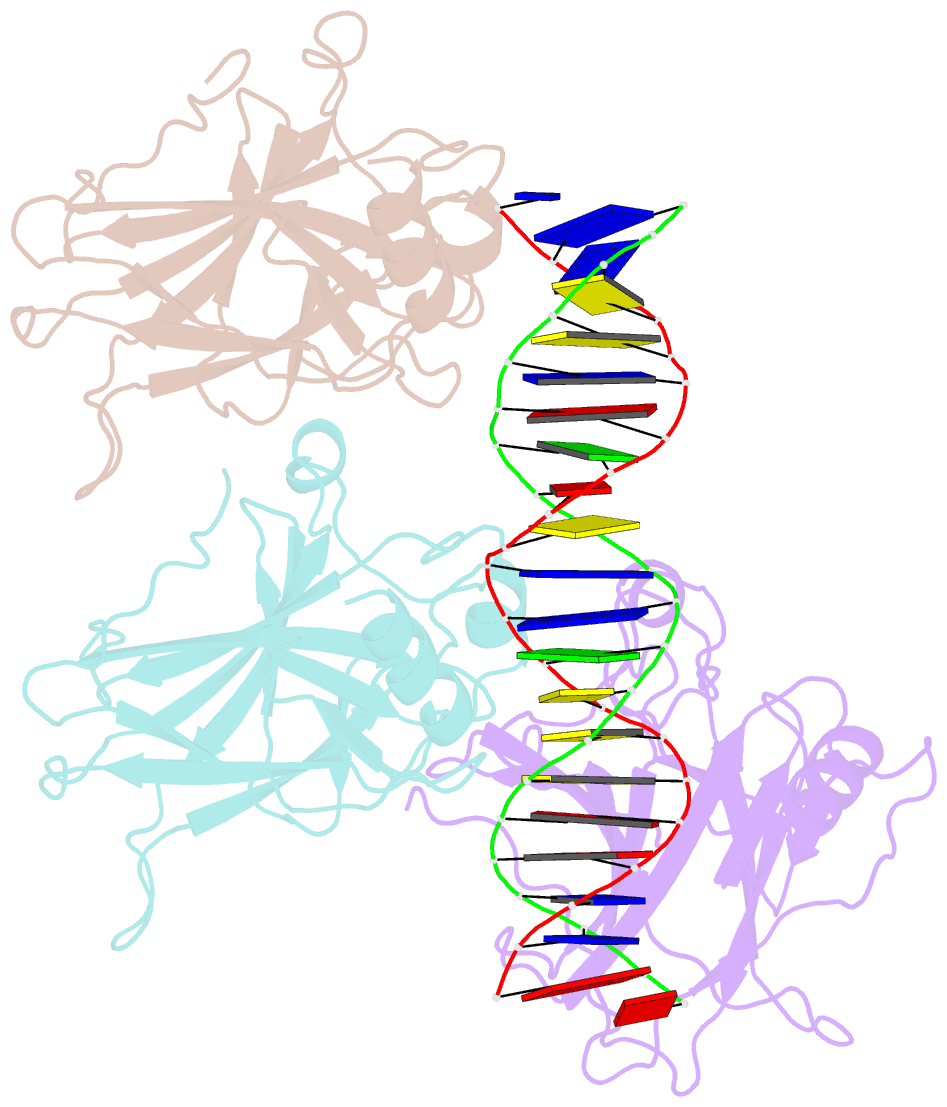
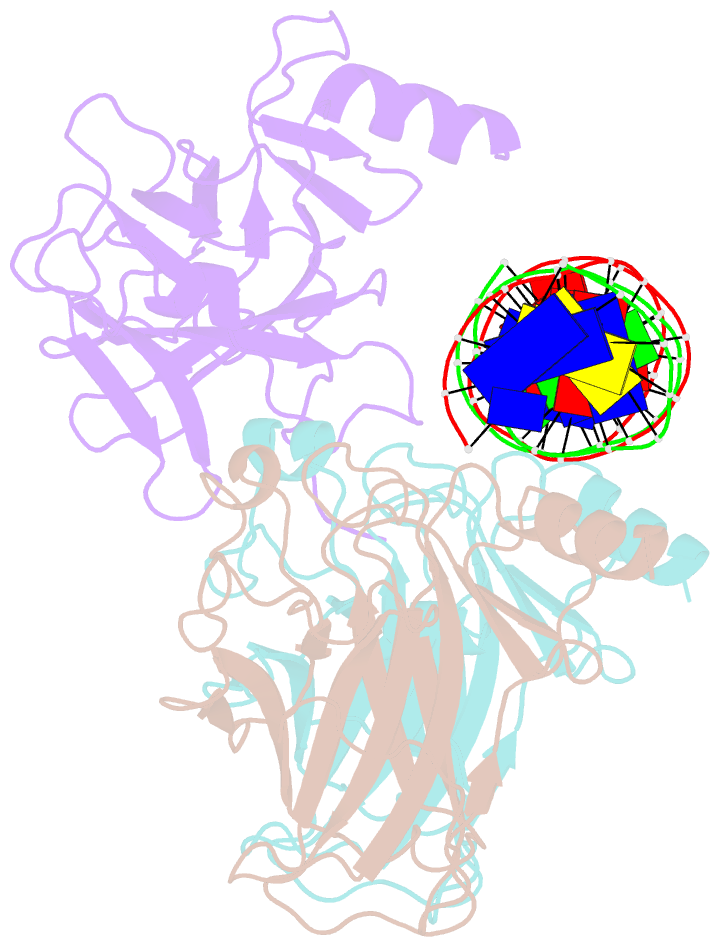
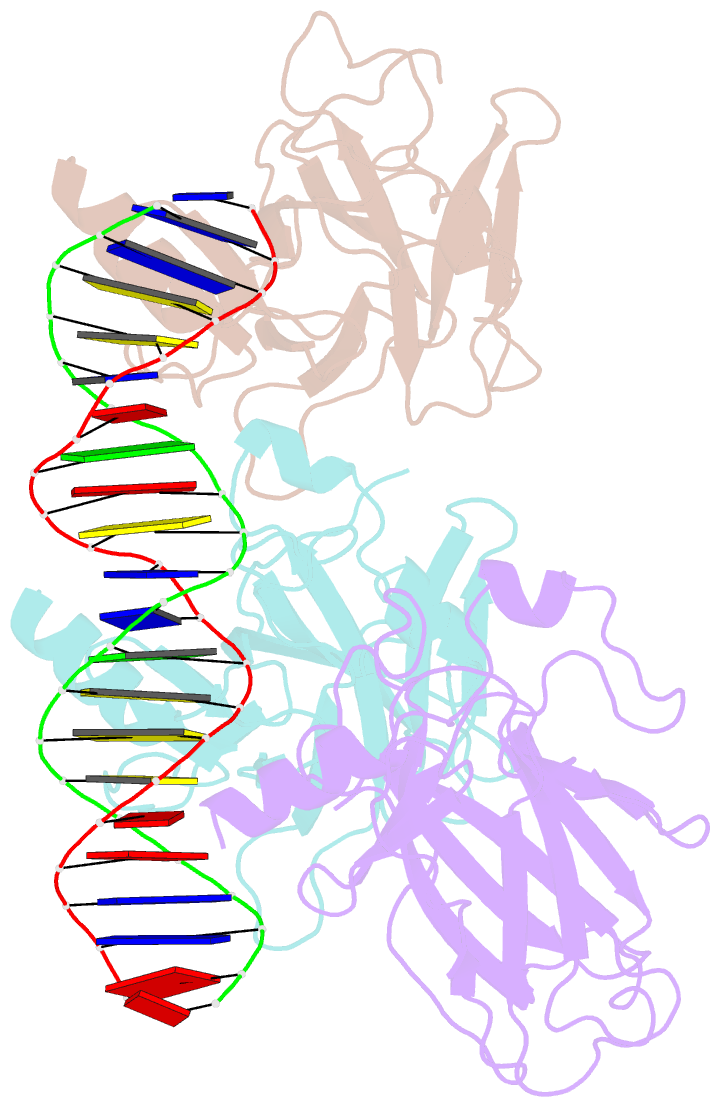
- Tumor suppressor p53 complexed with DNA
- Reference
- Cho Y, Gorina S, Jeffrey PD, Pavletich NP (1994): "Crystal structure of a p53 tumor suppressor-DNA complex: understanding tumorigenic mutations." Science, 265, 346-355.
- Abstract
- Mutations in the p53 tumor suppressor are the most frequently observed genetic alterations in human cancer. The majority of the mutations occur in the core domain which contains the sequence-specific DNA binding activity of the p53 protein (residues 102-292), and they result in loss of DNA binding. The crystal structure of a complex containing the core domain of human p53 and a DNA binding site has been determined at 2.2 angstroms resolution and refined to a crystallographic R factor of 20.5 percent. The core domain structure consists of a beta sandwich that serves as a scaffold for two large loops and a loop-sheet-helix motif. The two loops, which are held together in part by a tetrahedrally coordinated zinc atom, and the loop-sheet-helix motif form the DNA binding surface of p53. Residues from the loop-sheet-helix motif interact in the major groove of the DNA, while an arginine from one of the two large loops interacts in the minor groove. The loops and the loop-sheet-helix motif consist of the conserved regions of the core domain and contain the majority of the p53 mutations identified in tumors. The structure supports the hypothesis that DNA binding is critical for the biological activity of p53, and provides a framework for understanding how mutations inactivate it.