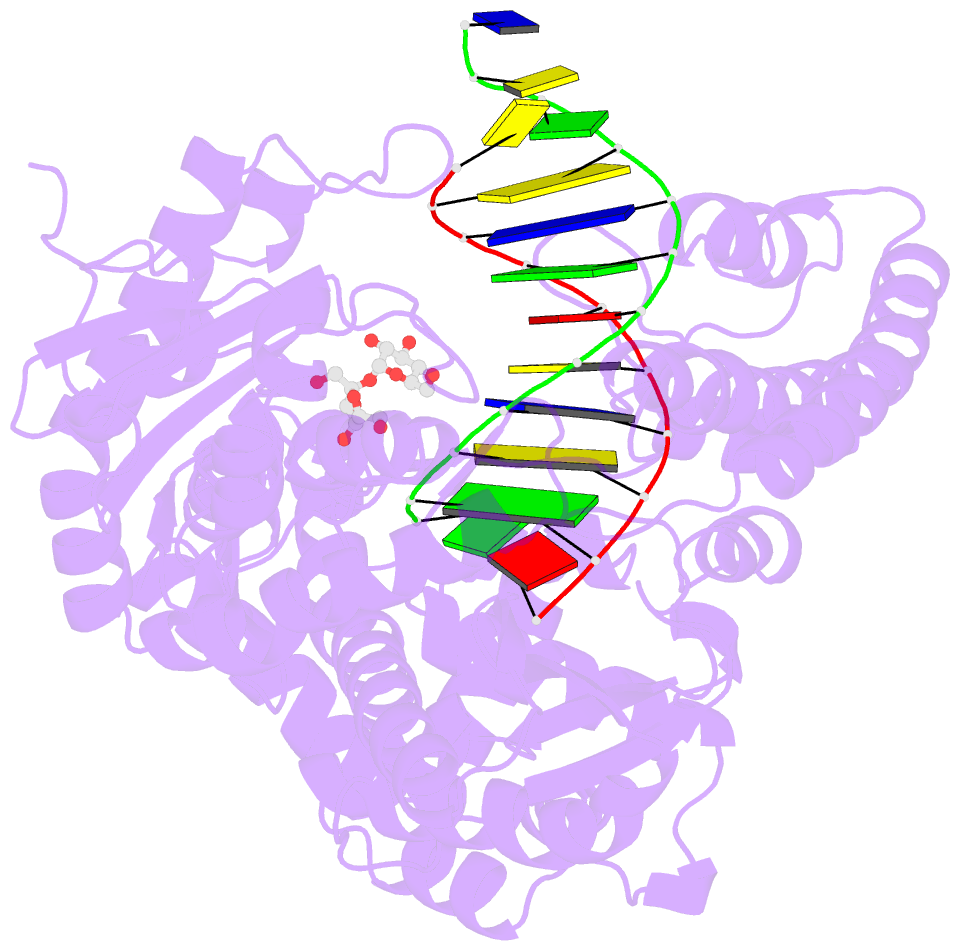
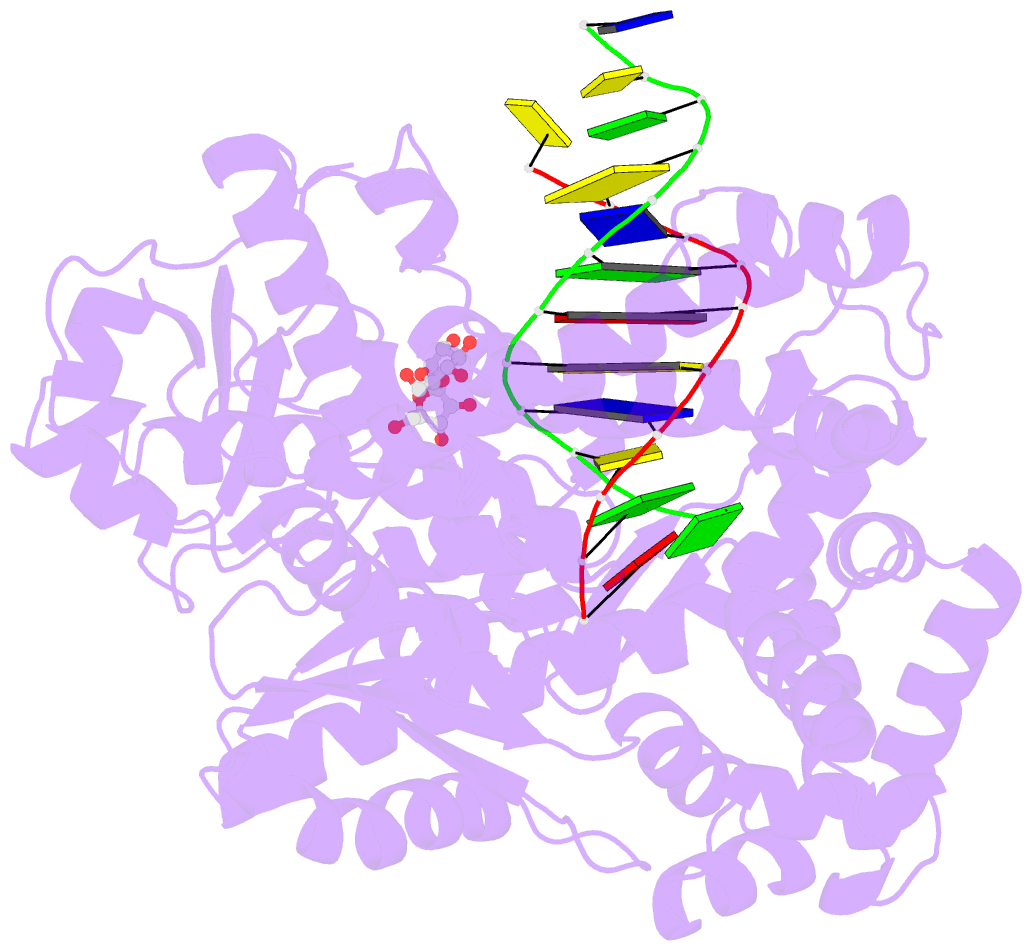
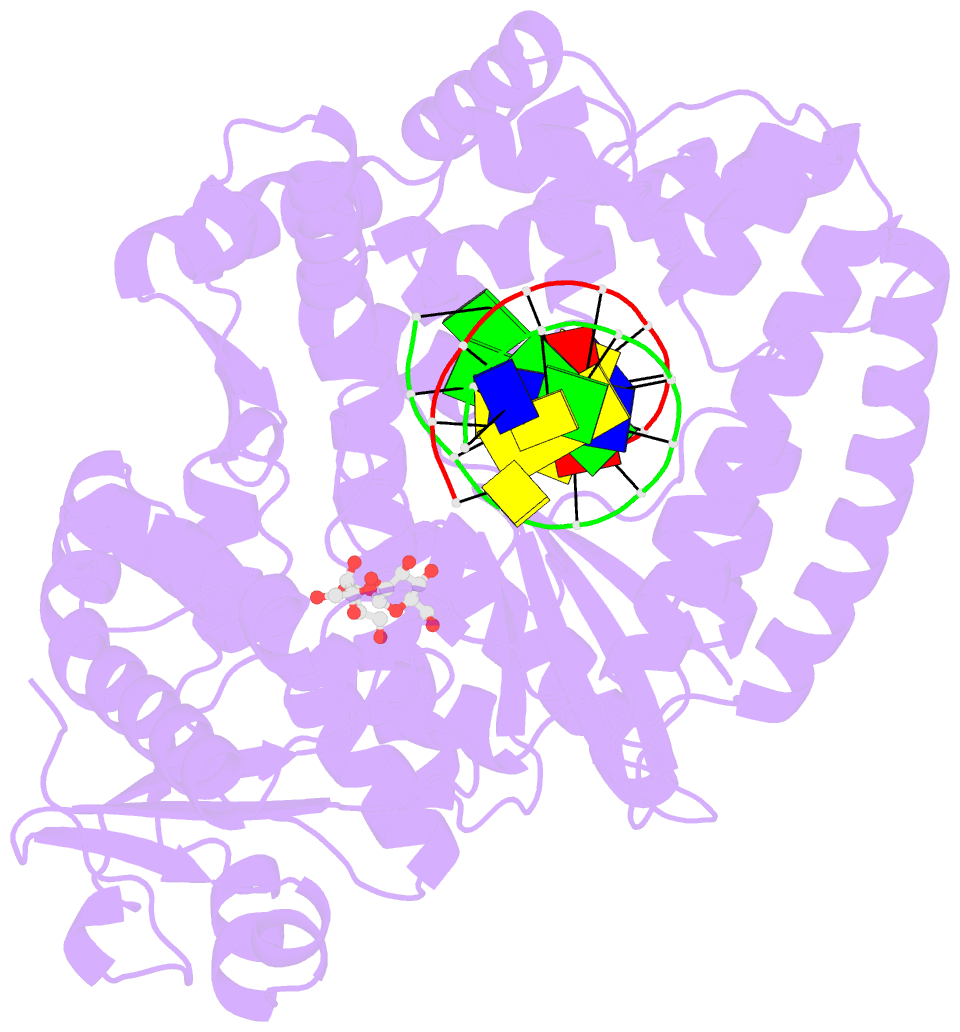
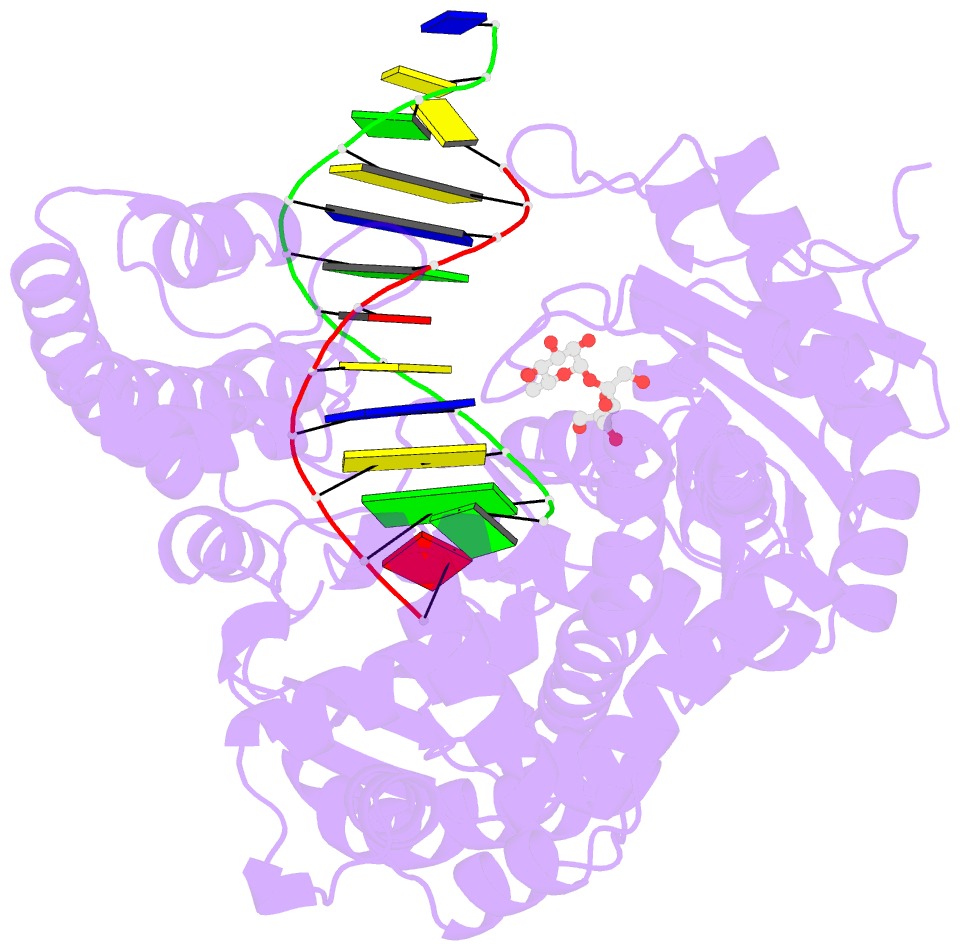
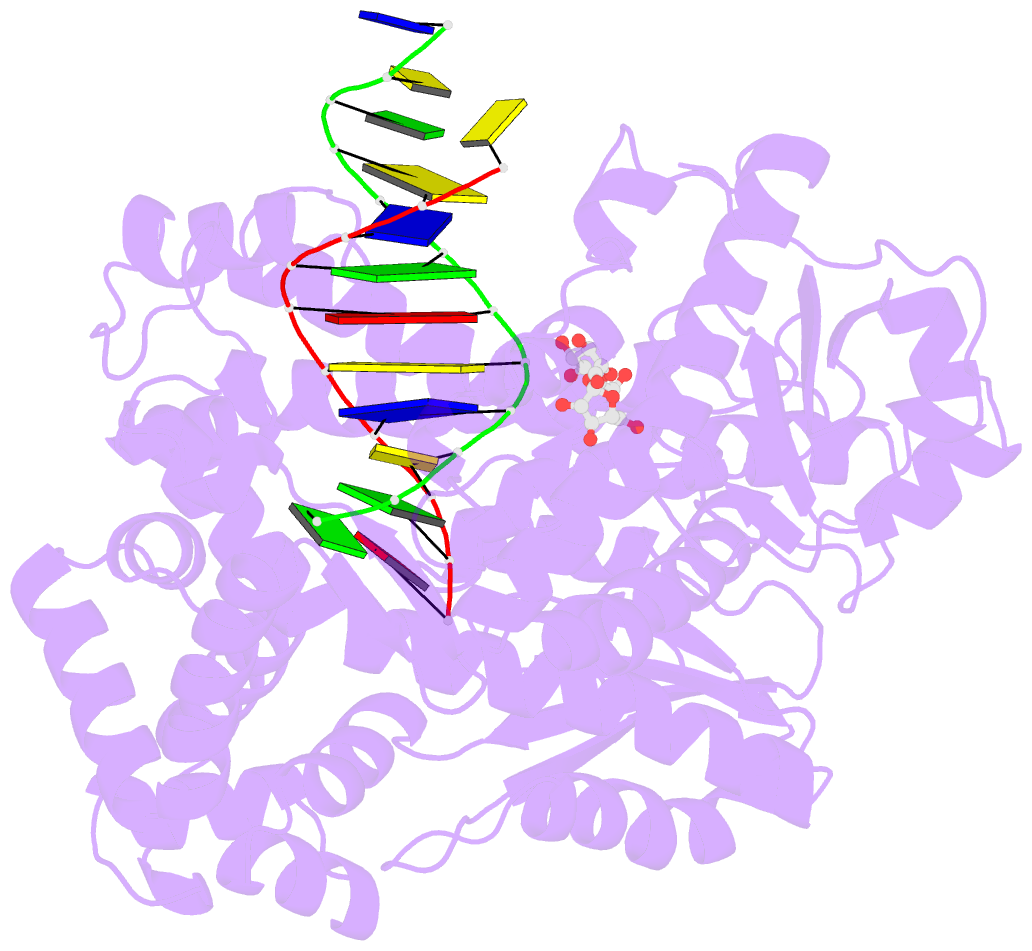
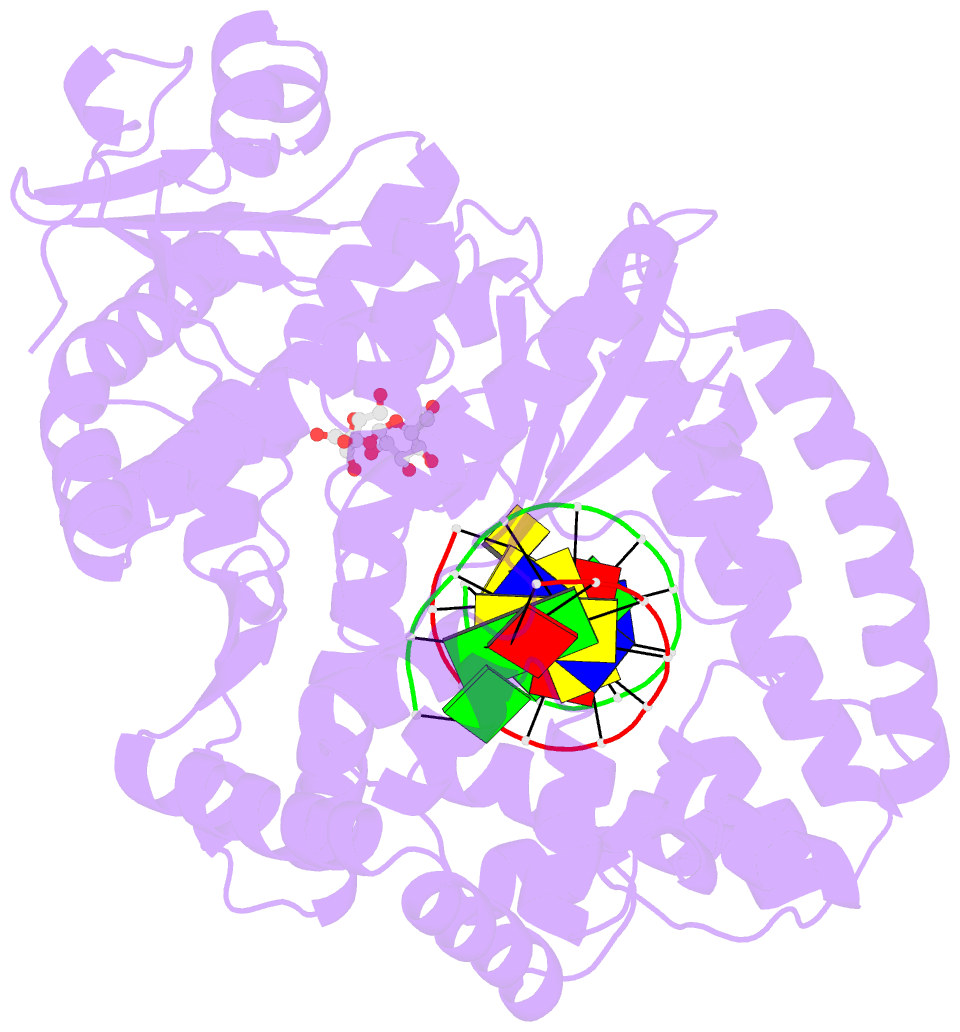
Summary information and primary citation
- PDB-id
- 1u49; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transferase-DNA
- Method
- X-ray (2.15 Å)
- Summary
- Adenine-8oxoguanine mismatch at the polymerase active site
- Reference
- Hsu GW, Ober M, Carell T, Beese LS (2004): "Error-prone replication of oxidatively damaged DNA by a high-fidelity DNA polymerase." Nature, 431, 217-221. doi: 10.1038/nature02908.
- Abstract
- Aerobic respiration generates reactive oxygen species that can damage guanine residues and lead to the production of 8-oxoguanine (8oxoG), the major mutagenic oxidative lesion in the genome. Oxidative damage is implicated in ageing and cancer, and its prevalence presents a constant challenge to DNA polymerases that ensure accurate transmission of genomic information. When these polymerases encounter 8oxoG, they frequently catalyse misincorporation of adenine in preference to accurate incorporation of cytosine. This results in the propagation of G to T transversions, which are commonly observed somatic mutations associated with human cancers. Here, we present sequential snapshots of a high-fidelity DNA polymerase during both accurate and mutagenic replication of 8oxoG. Comparison of these crystal structures reveals that 8oxoG induces an inversion of the mismatch recognition mechanisms that normally proofread DNA, such that the 8oxoG.adenine mismatch mimics a cognate base pair whereas the 8oxoG.cytosine base pair behaves as a mismatch. These studies reveal a fundamental mechanism of error-prone replication and show how 8oxoG, and DNA lesions in general, can form mismatches that evade polymerase error-detection mechanisms, potentially leading to the stable incorporation of lethal mutations.