Summary information and primary citation
- PDB-id
- 1u63; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transcription-RNA
- Method
- X-ray (3.4 Å)
- Summary
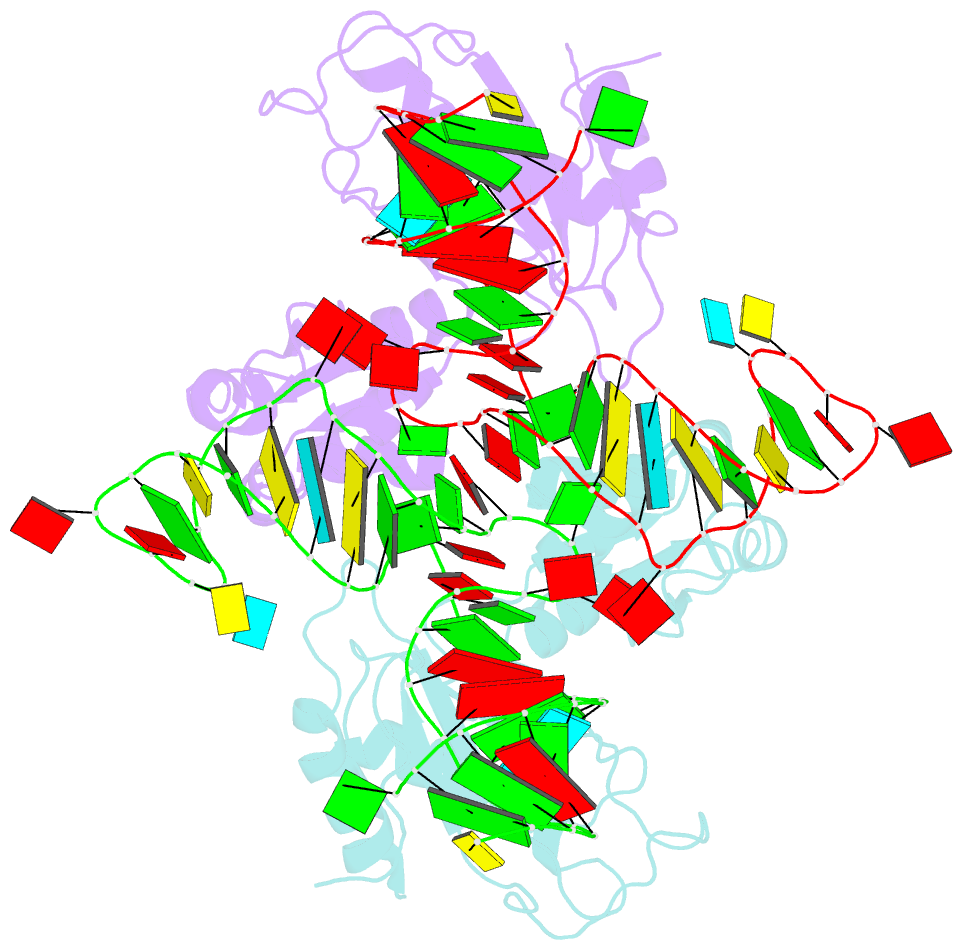
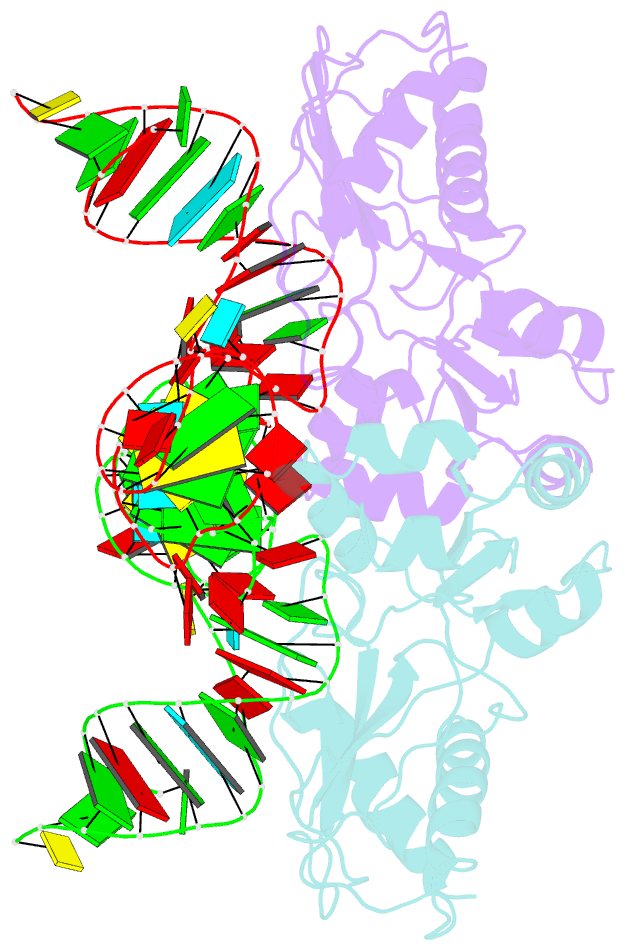
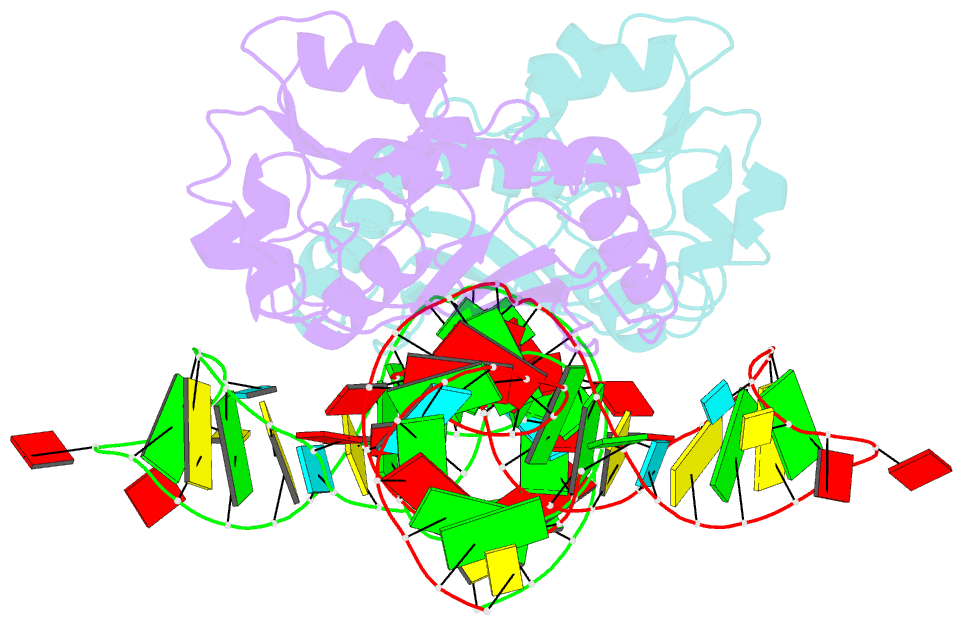
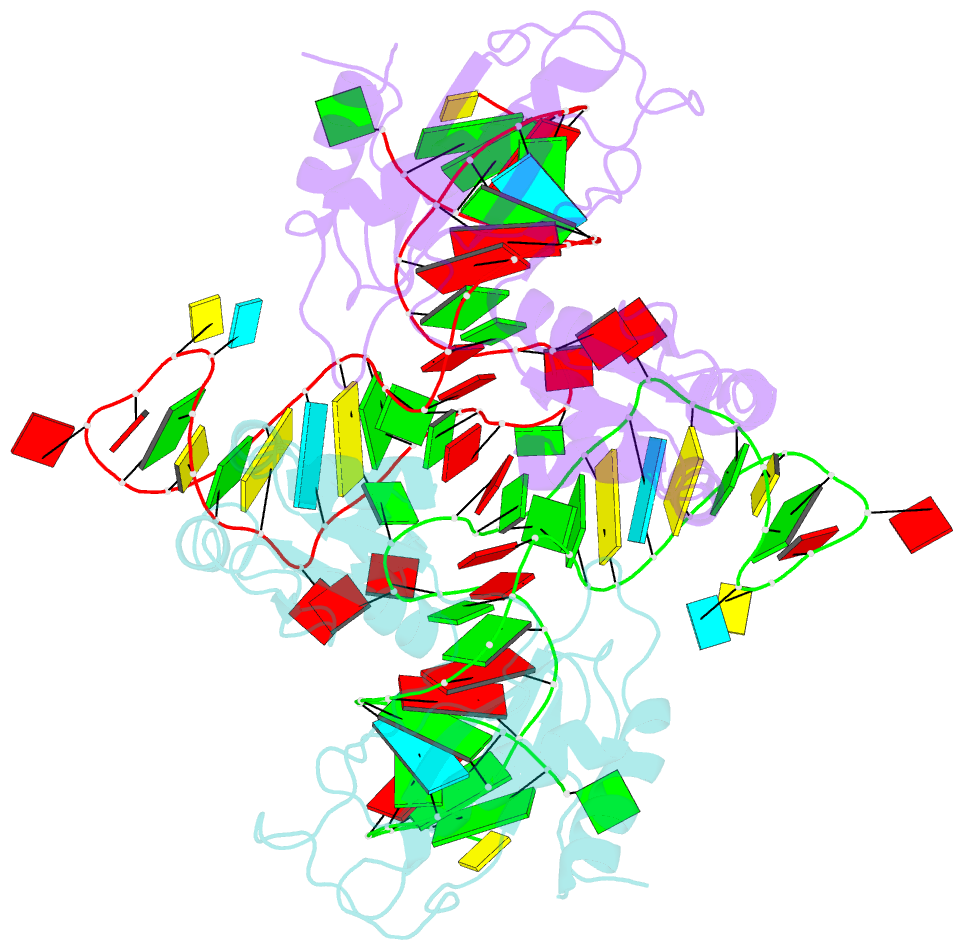
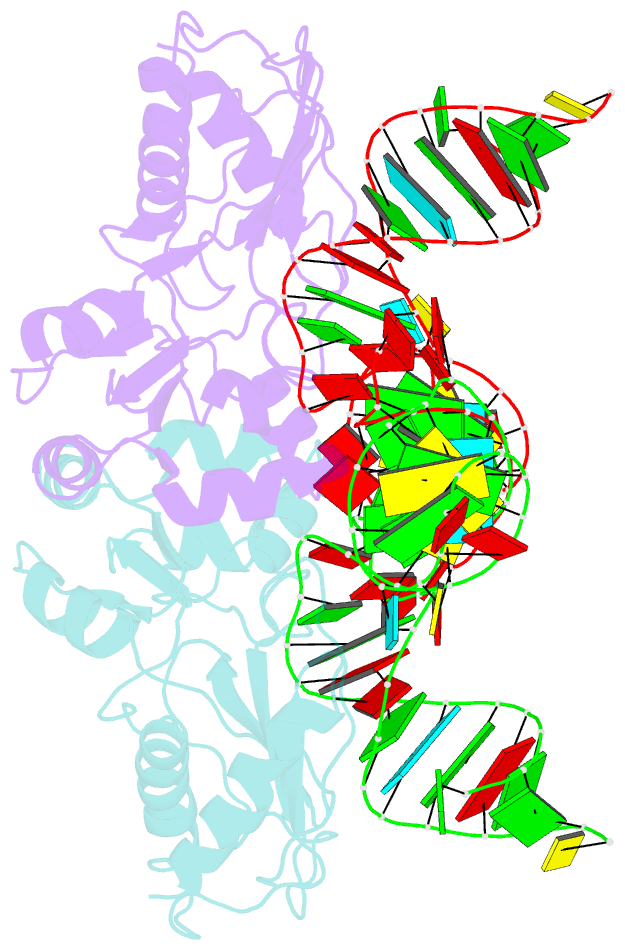
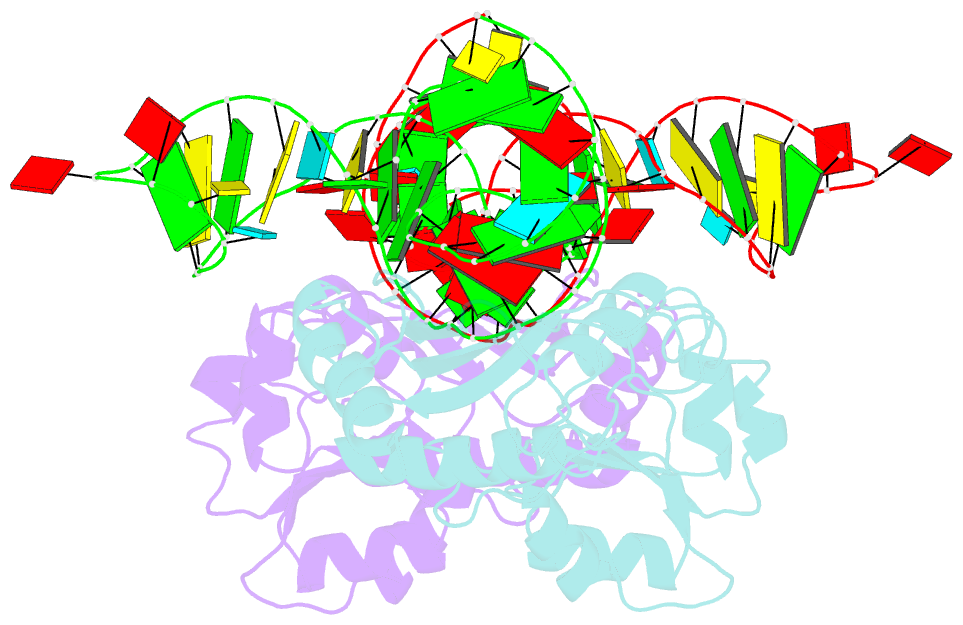
- The structure of a ribosomal protein l1-mrna complex
- Reference
- Nevskaya N, Tishchenko S, Gabdoulkhakov A, Nikonova E, Nikonov O, Nikulin A, Platonova O, Garber M, Nikonov S, Piendl W (2005): "Ribosomal protein L1 recognizes the same specific structural motif in its target sites on the autoregulatory mRNA and 23S rRNA." Nucleic Acids Res., 33, 478-485. doi: 10.1093/nar/gki194.
- Abstract
- The RNA-binding ability of ribosomal protein L1 is of profound interest since the protein has a dual function as a ribosomal protein binding rRNA and as a translational repressor binding its mRNA. Here, we report the crystal structure of ribosomal protein L1 in complex with a specific fragment of its mRNA and compare it with the structure of L1 in complex with a specific fragment of 23S rRNA determined earlier. In both complexes, a strongly conserved RNA structural motif is involved in L1 binding through a conserved network of RNA-protein H-bonds inaccessible to the solvent. These interactions should be responsible for specific recognition between the protein and RNA. A large number of additional non-conserved RNA-protein H-bonds stabilizes both complexes. The added contribution of these non-conserved H-bonds makes the ribosomal complex much more stable than the regulatory one.