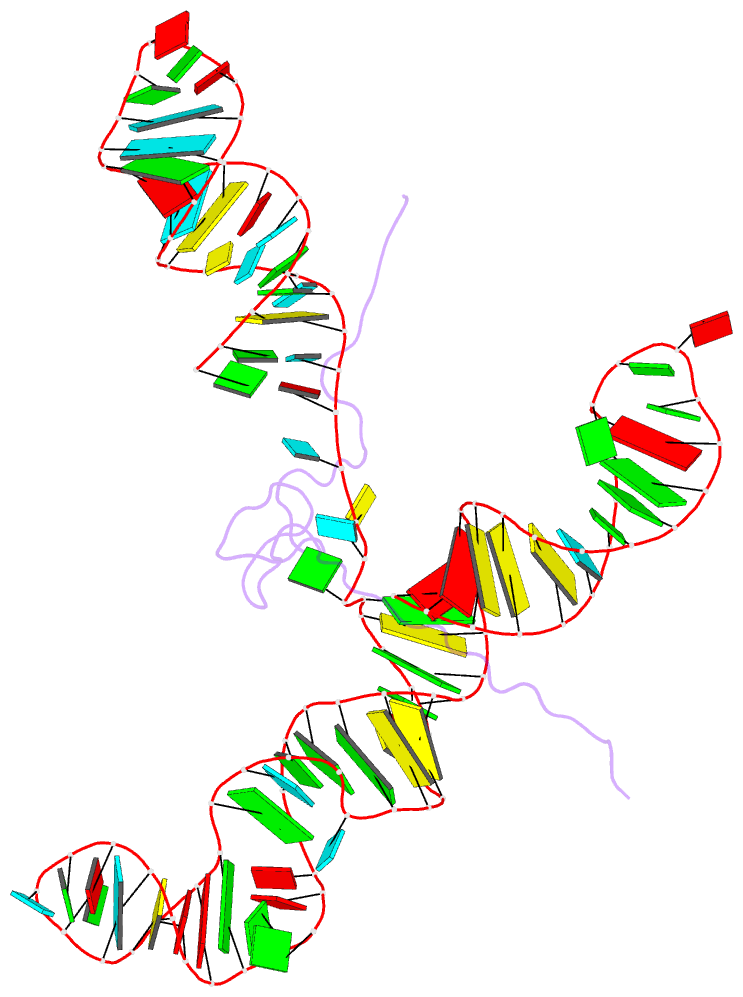
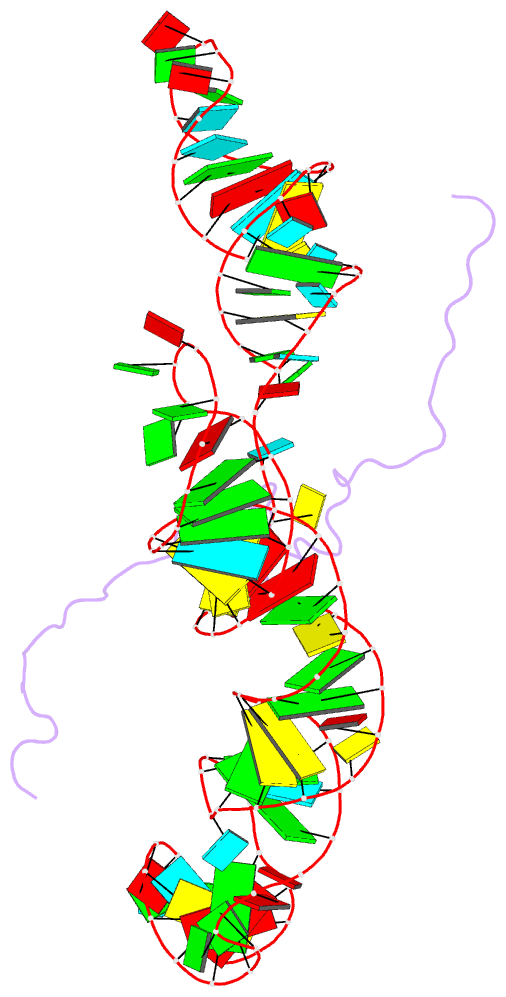
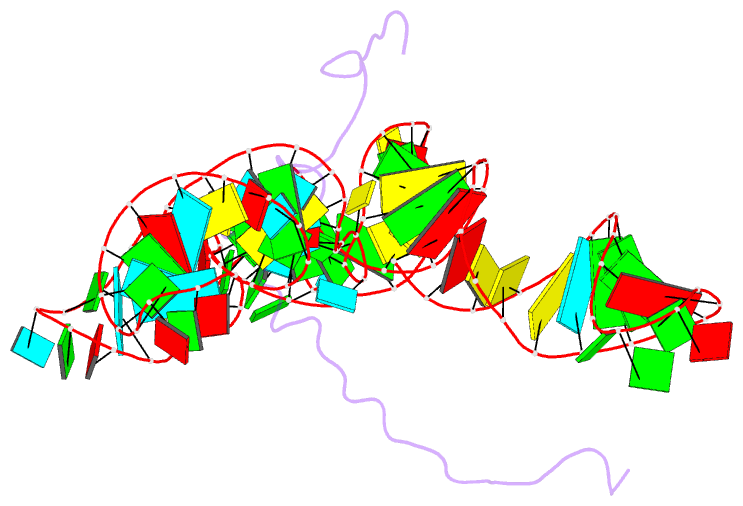
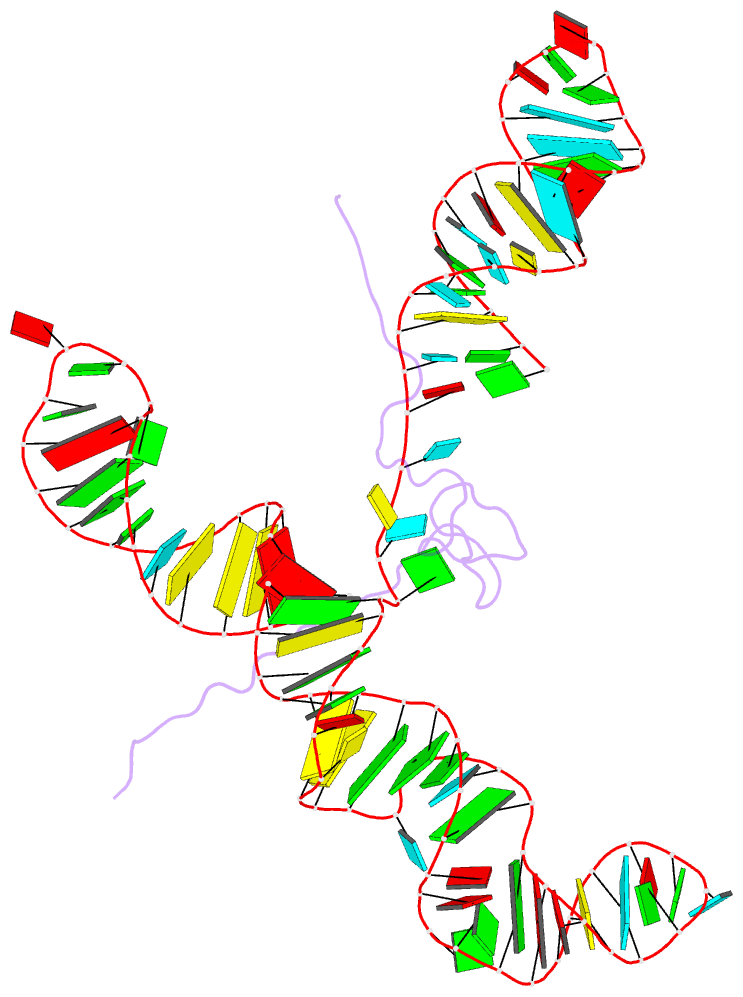
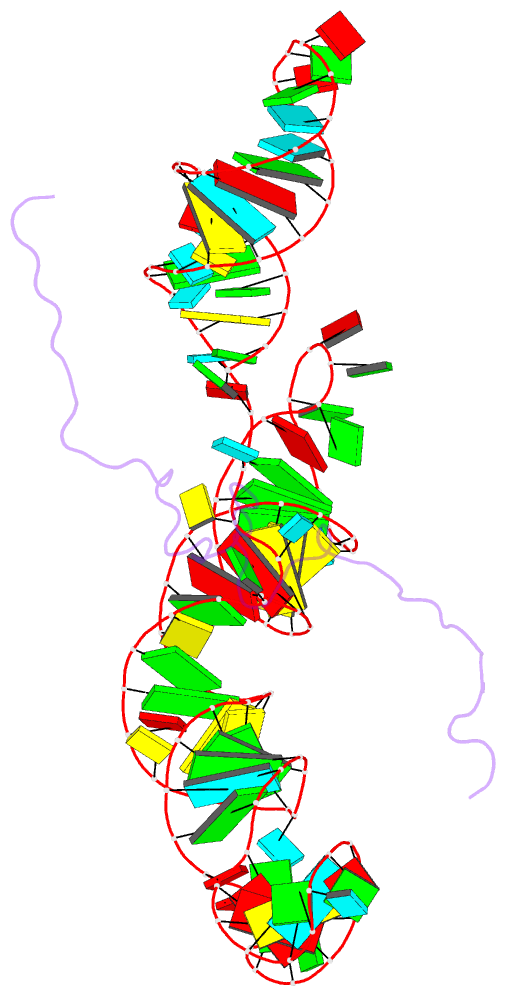
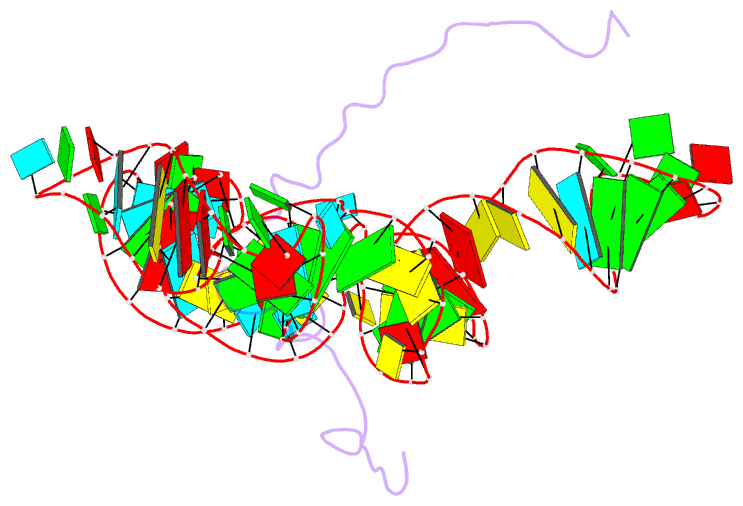
Summary information and primary citation
- PDB-id
- 1u6p; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- viral protein-RNA
- Method
- NMR
- Summary
- NMR structure of the mlv encapsidation signal bound to the nucleocapsid protein
- Reference
- D'Souza V, Summers MF (2004): "Structural basis for packaging the dimeric genome of Moloney murine leukaemia virus." Nature, 431, 586-590. doi: 10.1038/nature02944.
- Abstract
- All retroviruses specifically package two copies of their genomes during virus assembly, a requirement for strand-transfer-mediated recombination during reverse transcription. Genomic RNA exists in virions as dimers, and the overlap of RNA elements that promote dimerization and encapsidation suggests that these processes may be coupled. Both processes are mediated by the nucleocapsid domain (NC) of the retroviral Gag polyprotein. Here we show that dimerization-induced register shifts in base pairing within the Psi-RNA packaging signal of Moloney murine leukaemia virus (MoMuLV) expose conserved UCUG elements that bind NC with high affinity (dissociation constant = 75 +/- 12 nM). These elements are base-paired and do not bind NC in the monomeric RNA. The structure of the NC complex with a 101-nucleotide 'core encapsidation' segment of the MoMuLV Psi site reveals a network of interactions that promote sequence- and structure-specific binding by NC's single CCHC zinc knuckle. Our findings support a structural RNA switch mechanism for genome encapsidation, in which protein binding sites are sequestered by base pairing in the monomeric RNA and become exposed upon dimerization to promote packaging of a diploid genome.