Summary information and primary citation
- PDB-id
- 1u78; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- DNA binding protein-DNA
- Method
- X-ray (2.69 Å)
- Summary
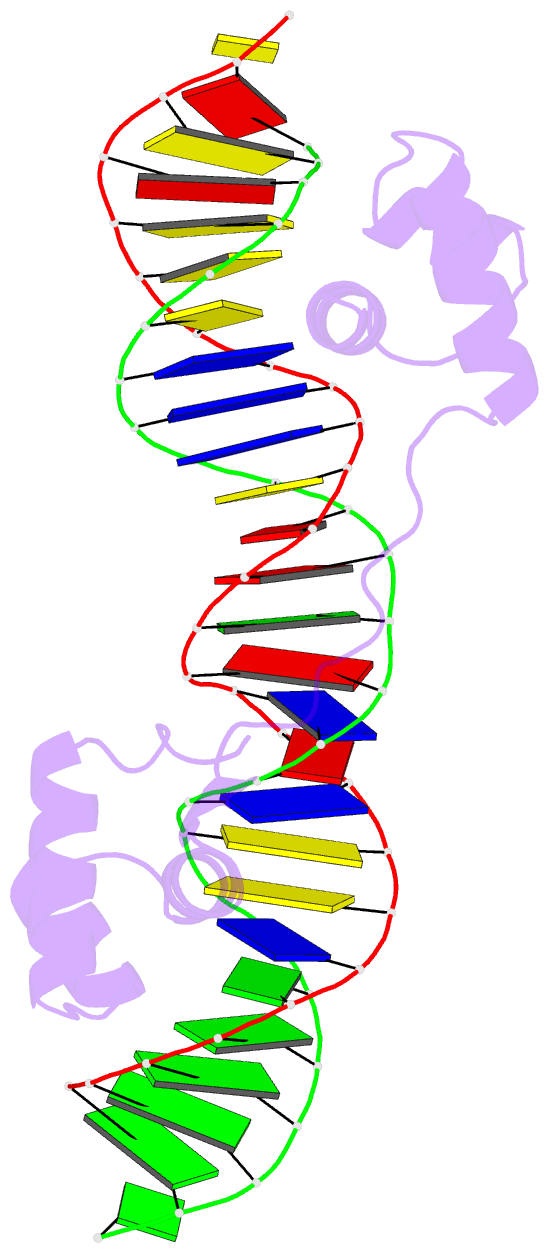
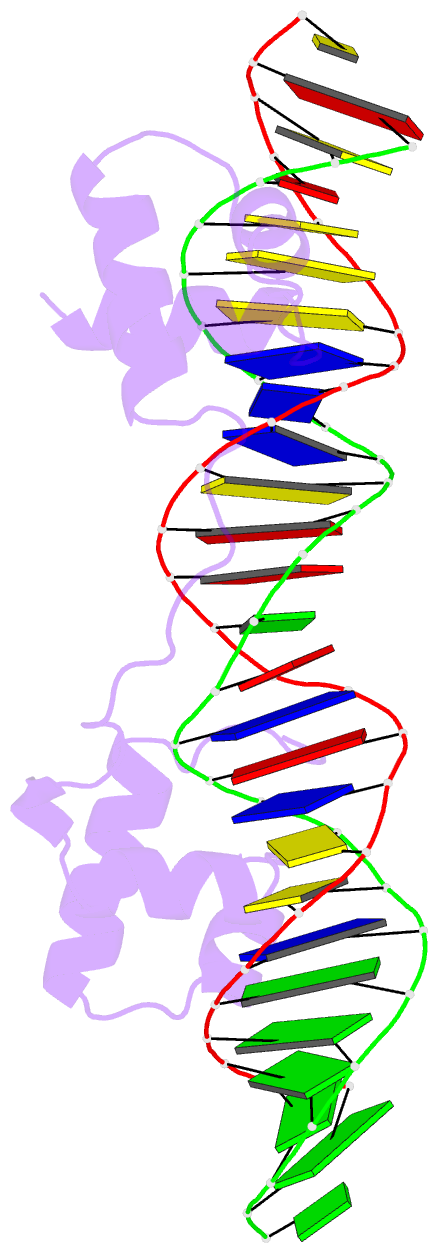
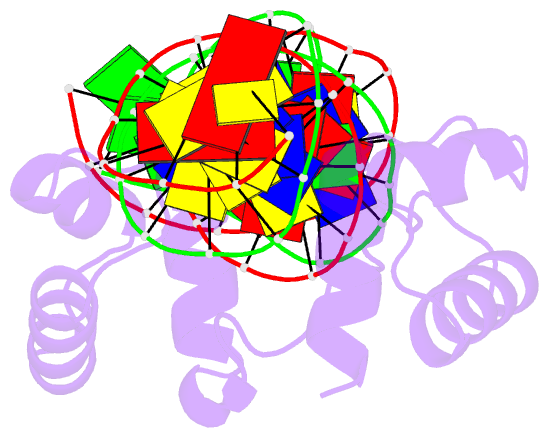
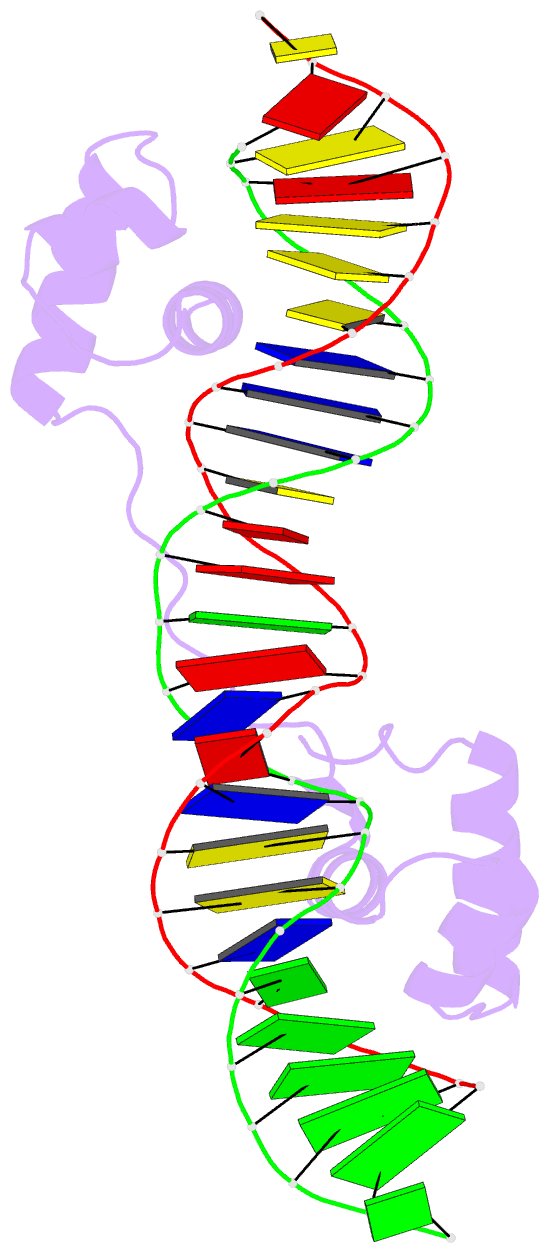
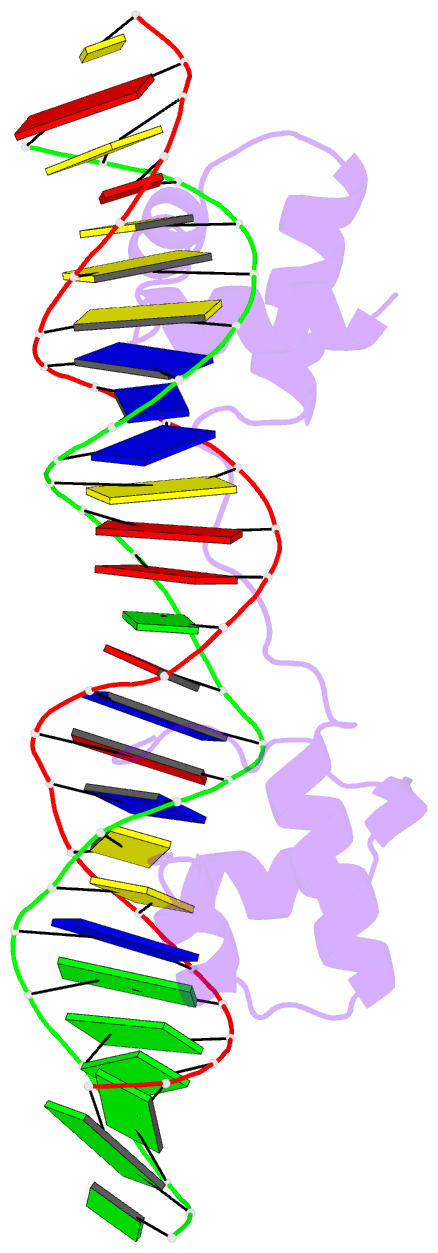
- Structure of the bipartite DNA-binding domain of tc3 transposase bound to transposon DNA
- Reference
- Watkins S, van Pouderoyen G, Sixma TK (2004): "Structural analysis of the bipartite DNA-binding domain of Tc3 transposase bound to transposon DNA." Nucleic Acids Res., 32, 4306-4312. doi: 10.1093/nar/gkh770.
- Abstract
- The bipartite DNA-binding domain of Tc3 transposase, Tc3A, was crystallized in complex with its transposon recognition sequence. In the structure the two DNA-binding domains form structurally related helix-turn-helix (HTH) motifs. They both bind to the major groove on a single DNA oligomer, separated by a linker that interacts closely with the minor groove. The structure resembles that of the transcription factor Pax6 DNA-binding domain, but the relative orientation of the HTH-domain is different. The DNA conformation is distorted, characterized by local narrowing of the minor groove and bends at both ends. The protein-DNA recognition takes place through base and backbone contacts, as well as shape-recognition of the distortions in the DNA. Charged interactions are primarily found in the N-terminal domain and the linker indicating that these may form the initial contact area. Two independent dimer interfaces could be relevant for bringing together transposon ends and for binding to a direct repeat site in the transposon end. In contrast to the Tn5 synaptic complex, the two Tc3A DNA-binding domains bind to a single Tc3 transposon end.