Summary information and primary citation
- PDB-id
- 1ua1; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transferase-DNA
- Method
- X-ray (2.0 Å)
- Summary
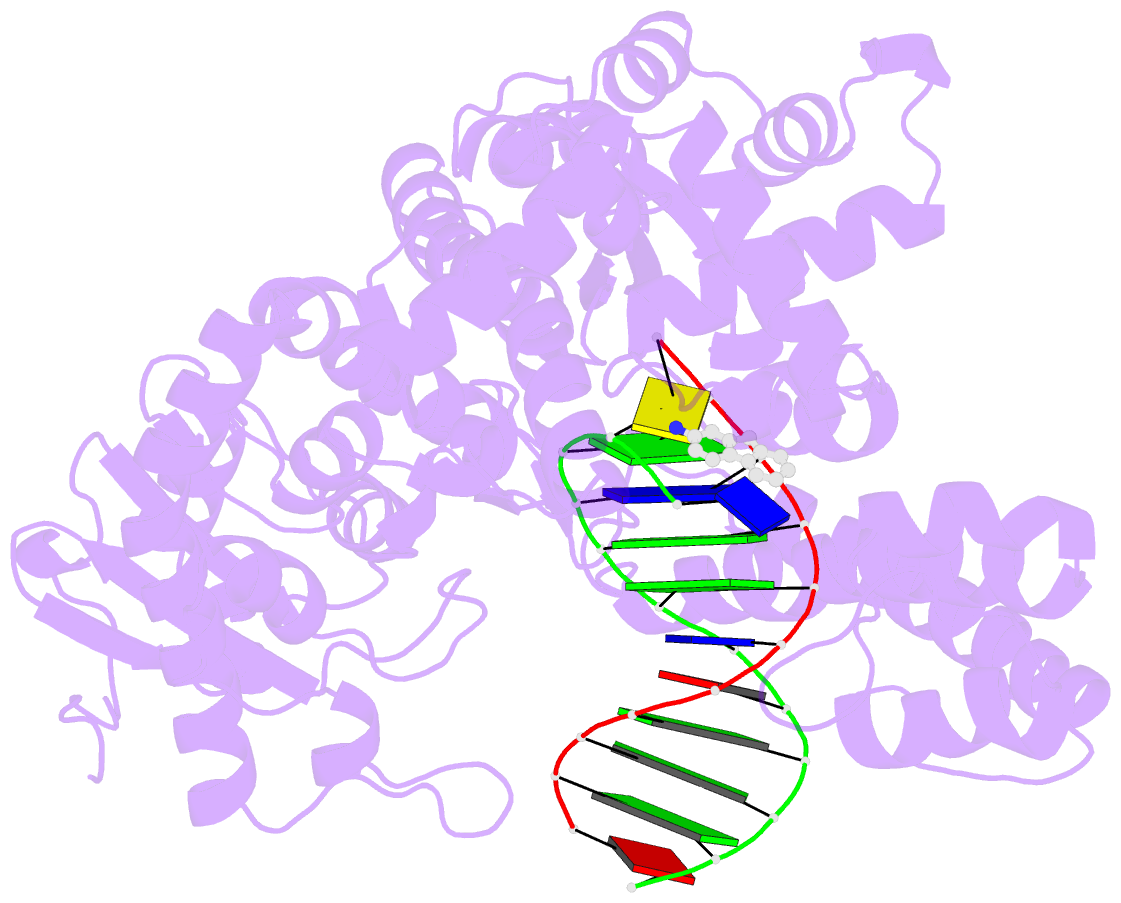
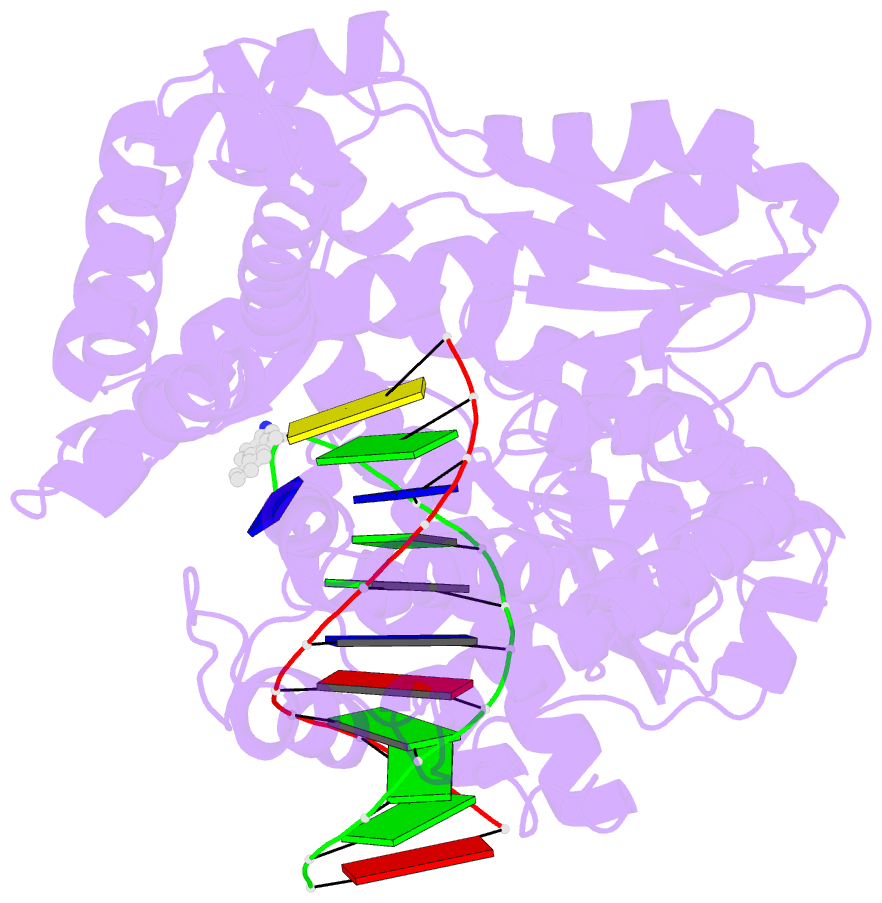
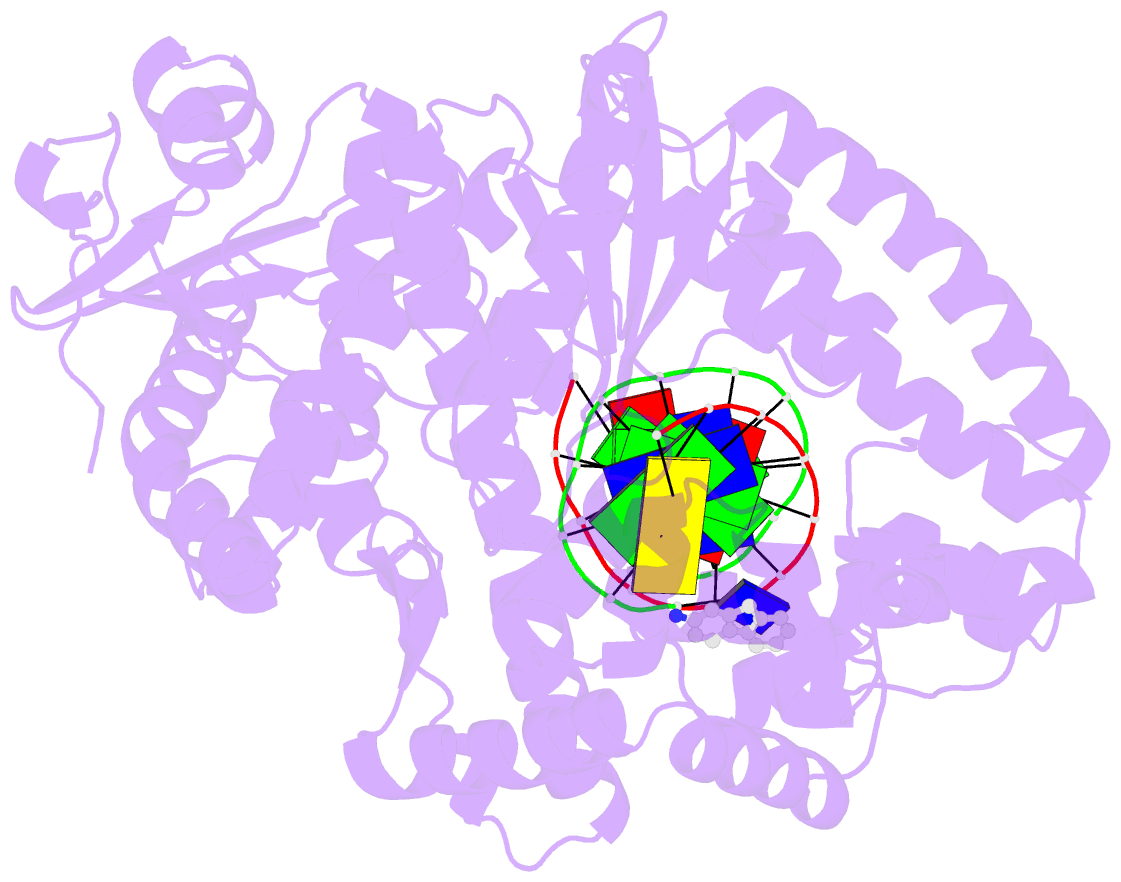
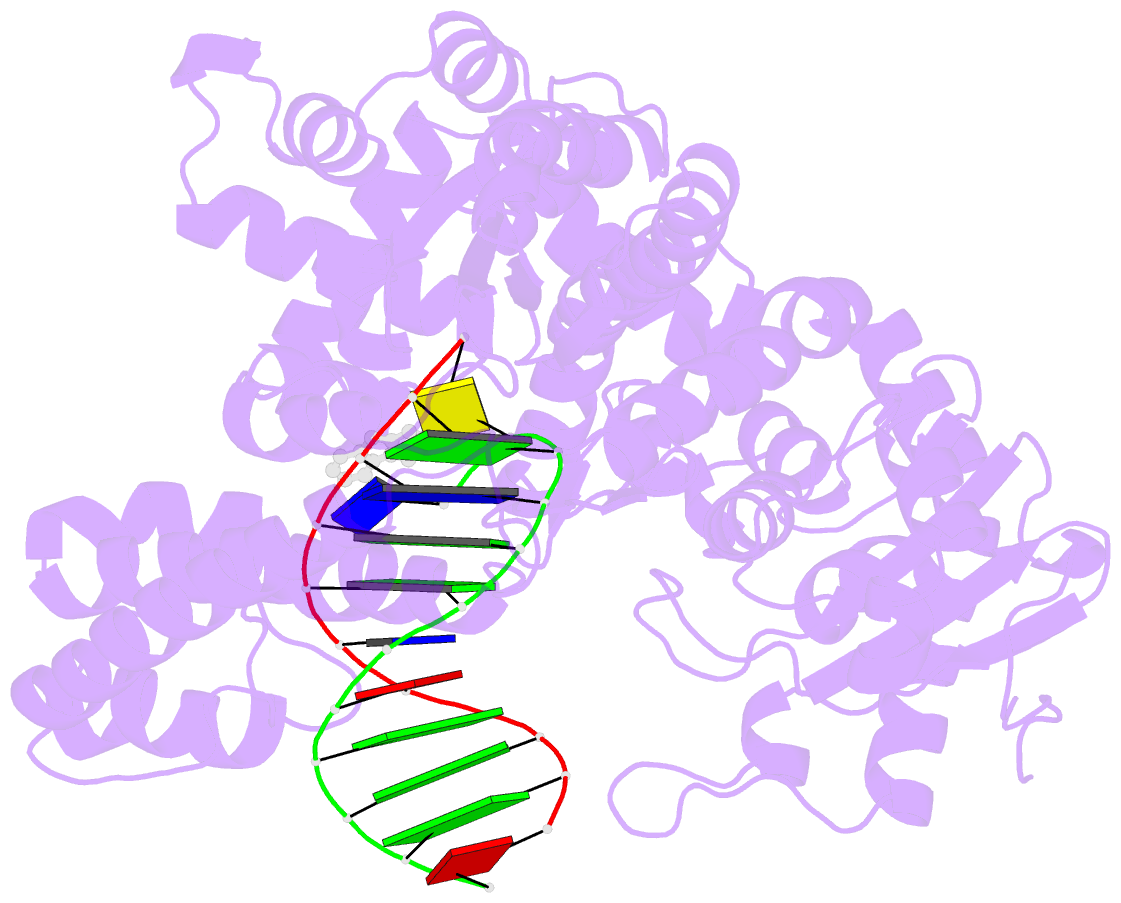
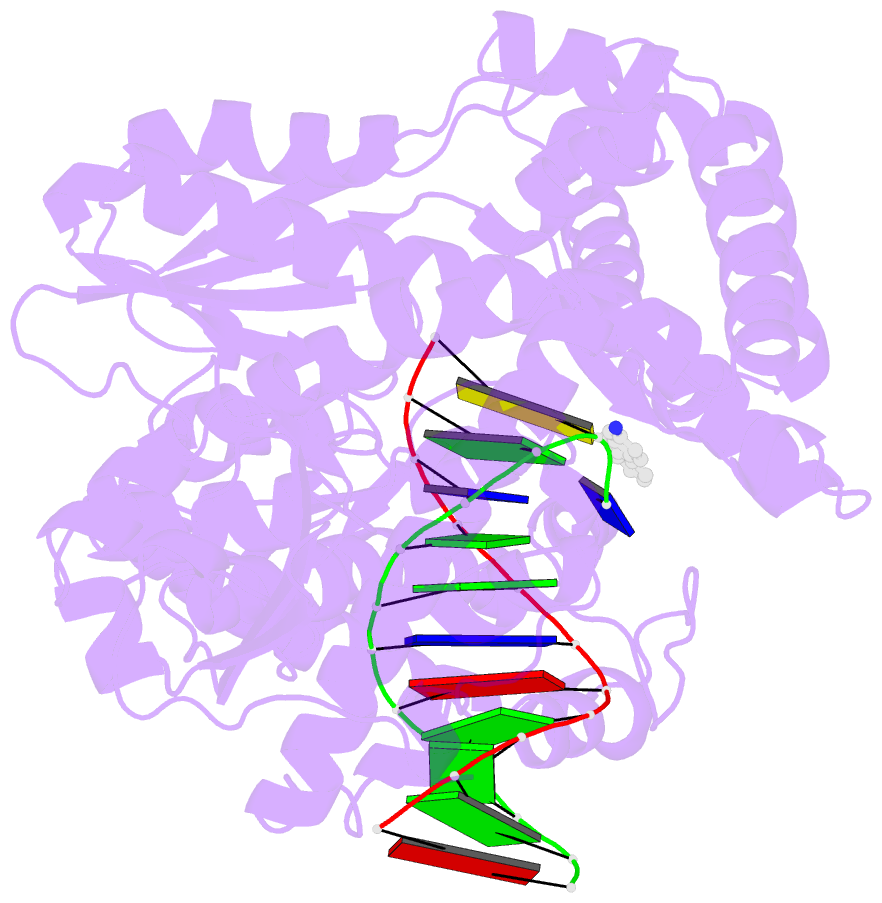
- Structure of aminofluorene adduct paired opposite cytosine at the polymerase active site.
- Reference
- Hsu GW, Kiefer JR, Becherel OJ, Fuchs RPP, Beese LS (2004): "Observing translesion synthesis of an aromatic amine DNA adduct by a high-fidelity DNA polymerase." J.Biol.Chem., 279, 50280-50285. doi: 10.1074/jbc.M409224200.
- Abstract
- Aromatic amines have been studied for more than a half-century as model carcinogens representing a class of chemicals that form bulky adducts to the C8 position of guanine in DNA. Among these guanine adducts, the N-(2'-deoxyguanosin-8-yl)-aminofluorene (G-AF) and N-2-(2'-deoxyguanosin-8-yl)-acetylaminofluorene (G-AAF) derivatives are the best studied. Although G-AF and G-AAF differ by only an acetyl group, they exert different effects on DNA replication by replicative and high-fidelity DNA polymerases. Translesion synthesis of G-AF is achieved with high-fidelity polymerases, whereas replication of G-AAF requires specialized bypass polymerases. Here we have presented structures of G-AF as it undergoes one round of accurate replication by a high-fidelity DNA polymerase. Nucleotide incorporation opposite G-AF is achieved in solution and in the crystal, revealing how the polymerase accommodates and replicates past G-AF, but not G-AAF. Like an unmodified guanine, G-AF adopts a conformation that allows it to form Watson-Crick hydrogen bonds with an opposing cytosine that results in protrusion of the bulky fluorene moiety into the major groove. Although incorporation opposite G-AF is observed, the C:G-AF base pair induces distortions to the polymerase active site that slow translesion synthesis.