Summary information and primary citation
- PDB-id
- 1ull; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- viral protein-RNA
- Method
- NMR
- Summary
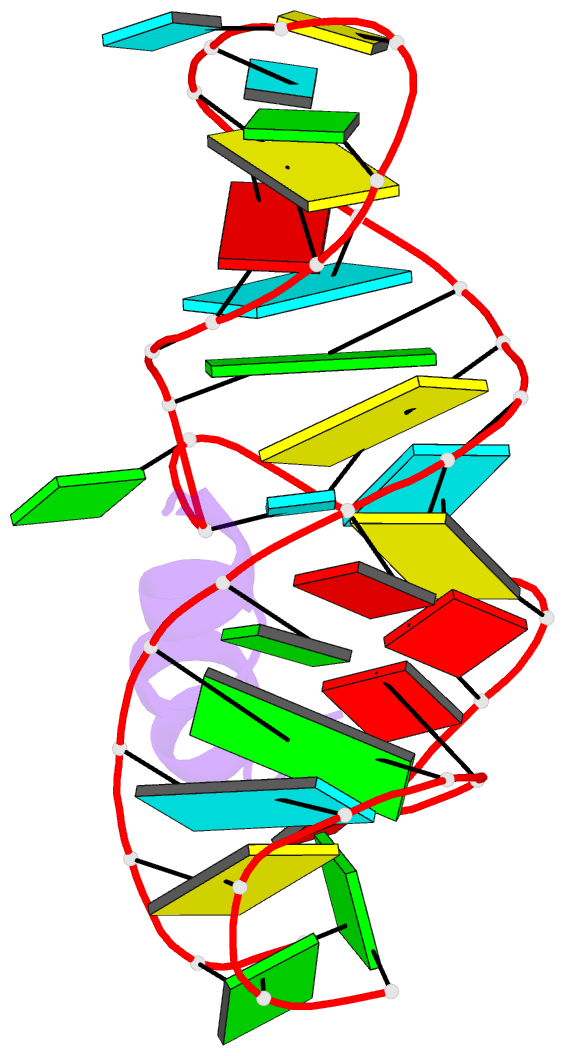
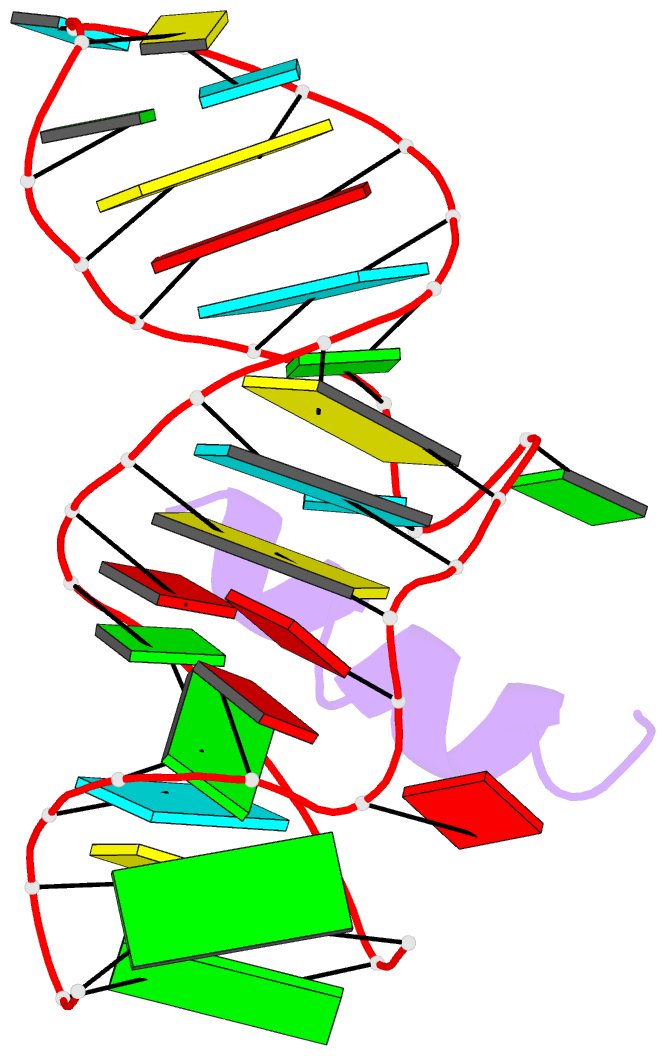
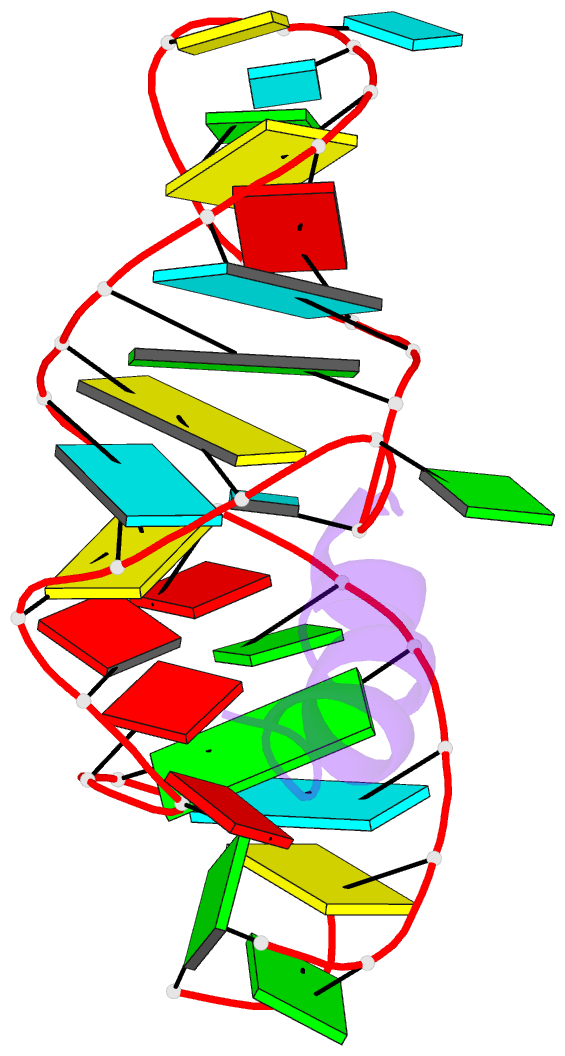
- RNA aptamer complexed with hiv-1 rev peptide, NMR, 7 structures
- Reference
- Ye X, Gorin A, Ellington AD, Patel DJ (1996): "Deep penetration of an alpha-helix into a widened RNA major groove in the HIV-1 rev peptide-RNA aptamer complex." Nat.Struct.Biol., 3, 1026-1033. doi: 10.1038/nsb1296-1026.
- Abstract
- A combined NMR-molecular dynamics approach has been applied to determine the solution structure of a HIV-1 17-mer rev peptide bound to its 35-mer high affinity RNA aptamer binding site. Complex formation involves adaptive binding with the alpha-helical arginine-rich basic rev peptide targeting a widened RNA major groove centred about adjacent G.A and reversed A.A mismatches. We have also identified a U AU triple in the aptamer complex with the Hoogsteen-paired uracil base sandwiched between two arginine side chains. The intermolecular contacts identified in the aptamer complex readily account for the consequences of peptide and RNA mutations, as well as the results of previous in vitro selection experiments. The details of molecular recognition associated with targeting by rev of its high affinity RNA binding sites open new opportunities for structure-based drug design strategies.