Summary information and primary citation
- PDB-id
- 1unm; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- DNA-antibiotic
- Method
- X-ray (2.0 Å)
- Summary
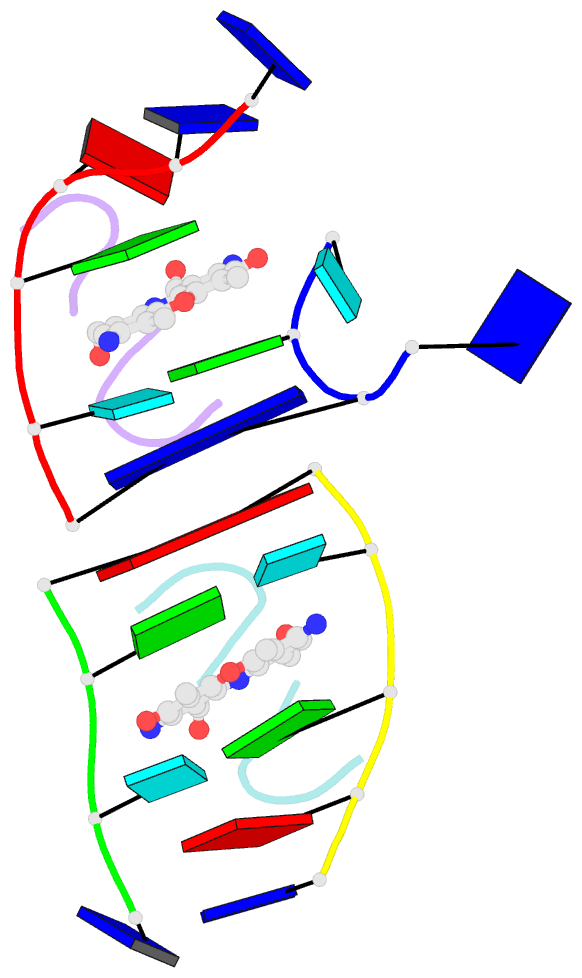
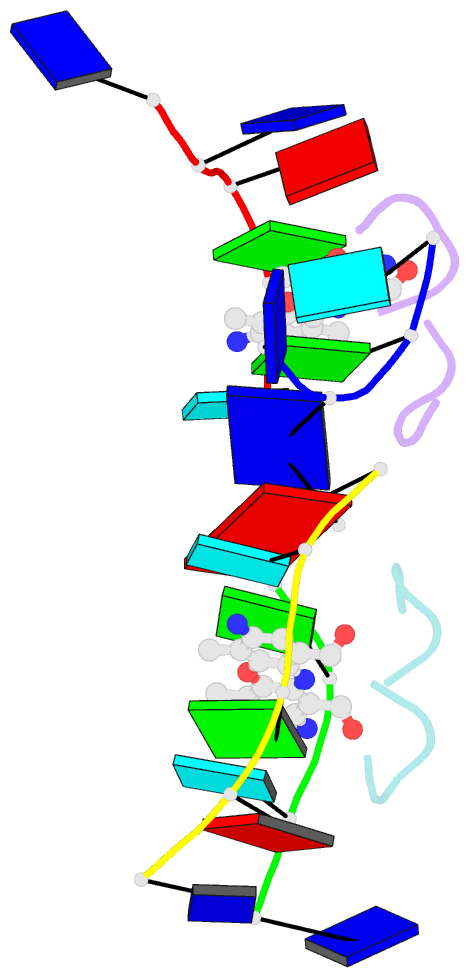
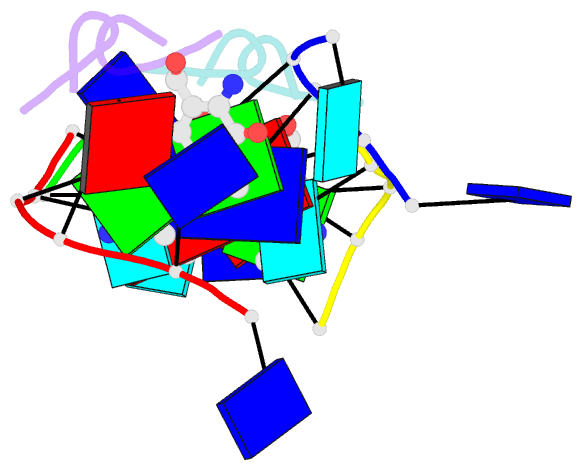
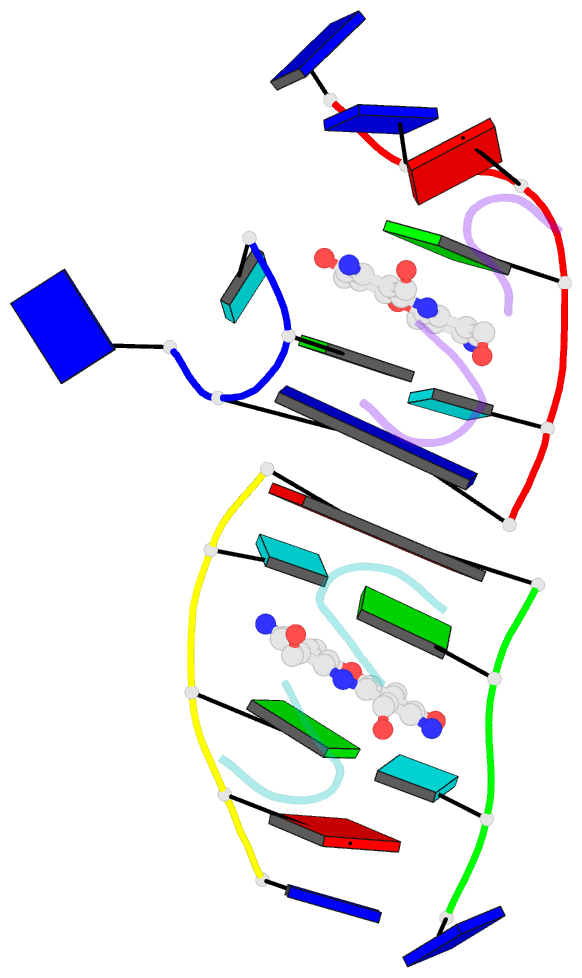
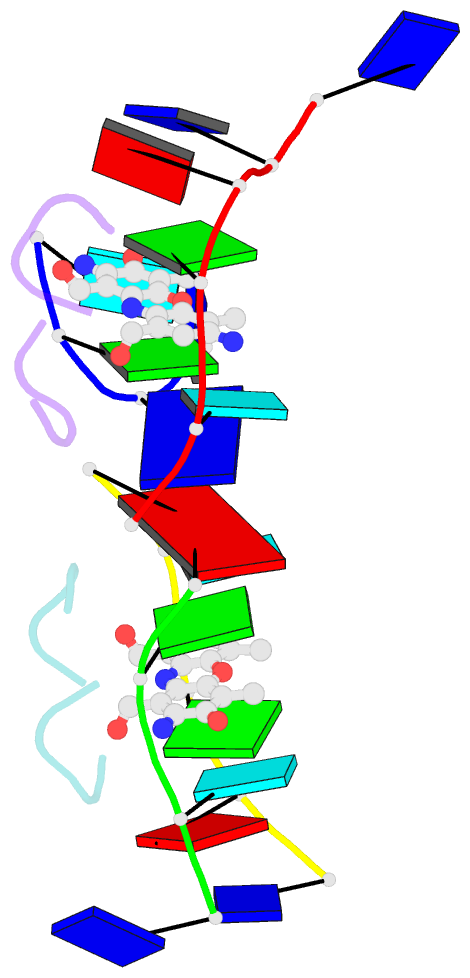
- Crystal structure of 7-aminoactinomycin d with non-complementary DNA
- Reference
- Alexopoulos EC, Jares-Erijman EA, Jovin TM, Klement R, Machinek R, Sheldrick GM, Uson I (2005): "Crystal and Solution Structures of 7-Amino-Actinomycin D Complexes with D(Ttagbrut), D(Ttagtt) and D(Tttagttt)." Acta Crystallogr.,Sect.D, 61, 407. doi: 10.1107/S090744490500082X.
- Abstract
- The formation of the complex of 7-amino-actinomycin D with potentially single-stranded DNA has been studied by X-ray crystallography in the solid state, by NMR in solution and by molecular modelling. The crystal structures of the complex with 5'-TTAG[Br(5)U]T-3' provide interesting examples of MAD phasing in which the dispersive component of the MAD signal was almost certainly enhanced by radiation damage. The trigonal and orthorhombic crystal modifications both contain antibiotic molecules and DNA strands in the form of a 2:4 complex: in the orthorhombic form there is one such complex in the asymmetric unit, while in the trigonal structure there are four. In both structures the phenoxazone ring of the first drug intercalates between a BrU-G (analogous to T-G) wobble pair and a G-T pair where the T is part of a symmetry-related molecule. The chromophore of the second actinomycin intercalates between the BrU-G and G-BrU wobble pairs of the partially paired third and fourth strands. The base stacking also involves (A*T)*T triplets and Watson-Crick A-T pairs and leads to similar complex three-dimensional networks in both structures, with looping-out of unpaired bases. Although the available NOE constraints of a solution containing the antibiotic and d(TTTAGTTT) strands in the ratio 1:1 are insufficient to determine the structure of the complex from the NMR data alone, they are consistent with the intercalation geometry observed in the crystal structure. Molecular-dynamics (MD) trajectories starting from the 1:2 complexes observed in the crystal showed that although the thymines flanking the d(AGT) core are rather flexible and the G-T pairing is not permanently preserved, both strands remain bound to the actinomycin by strong interactions between it and the guanines between which it is sandwiched. Similar strong binding (hemi-intercalation) of the actinomycin to a single guanine was observed in the MD trajectories of a 1:1 complex. The dominant interaction is between the antibiotic and guanine, but the complexes are stabilized further by promiscuous base-pairing.