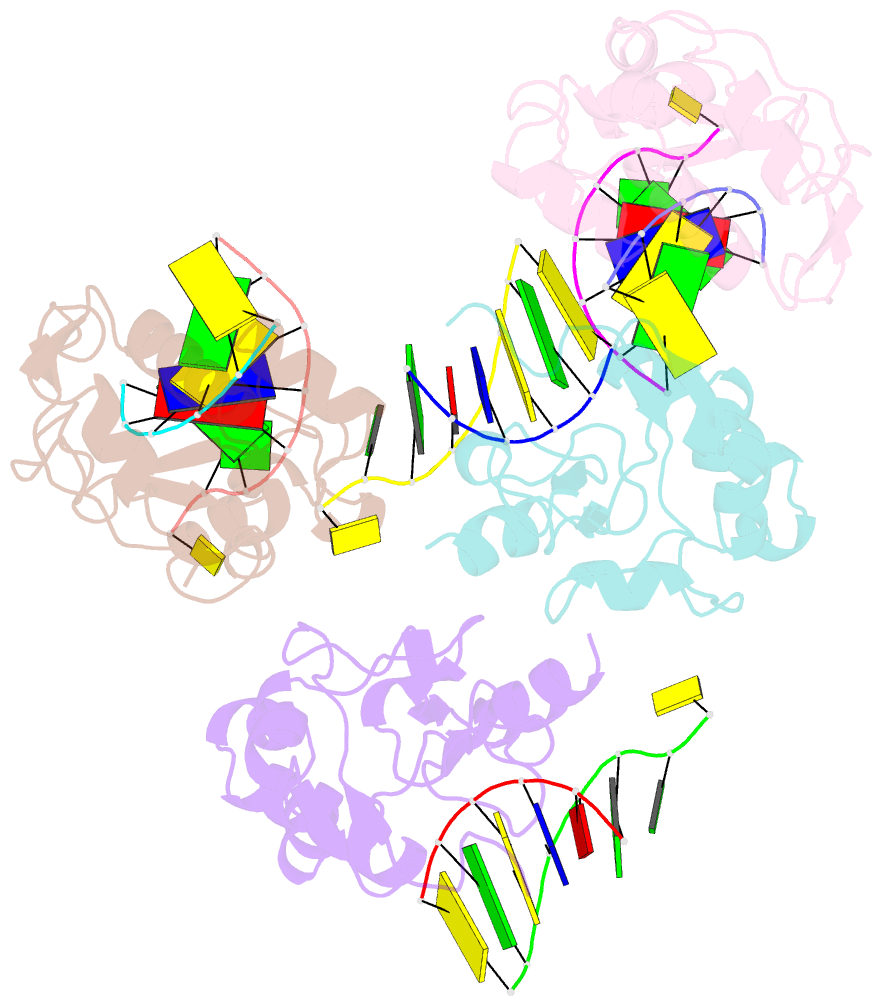
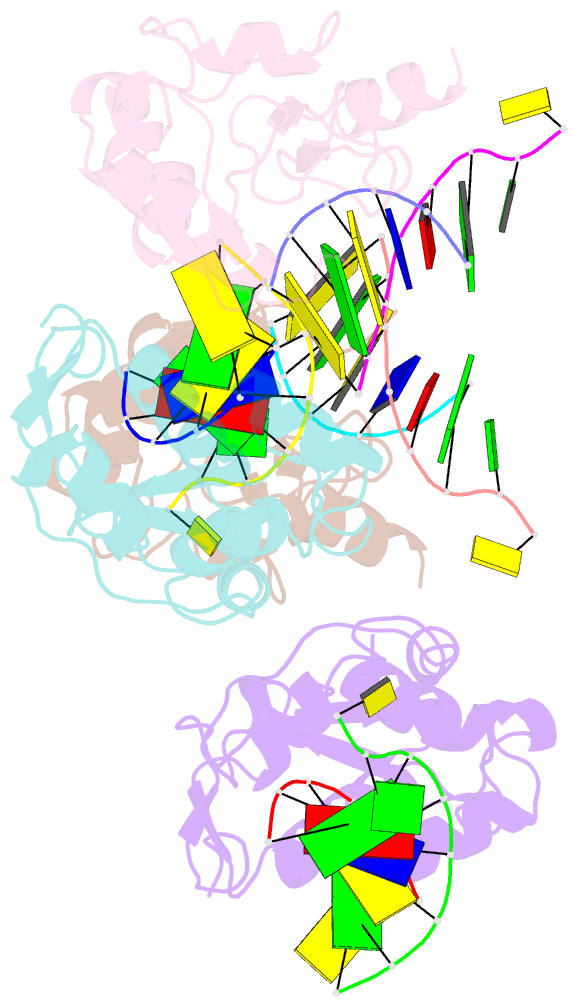
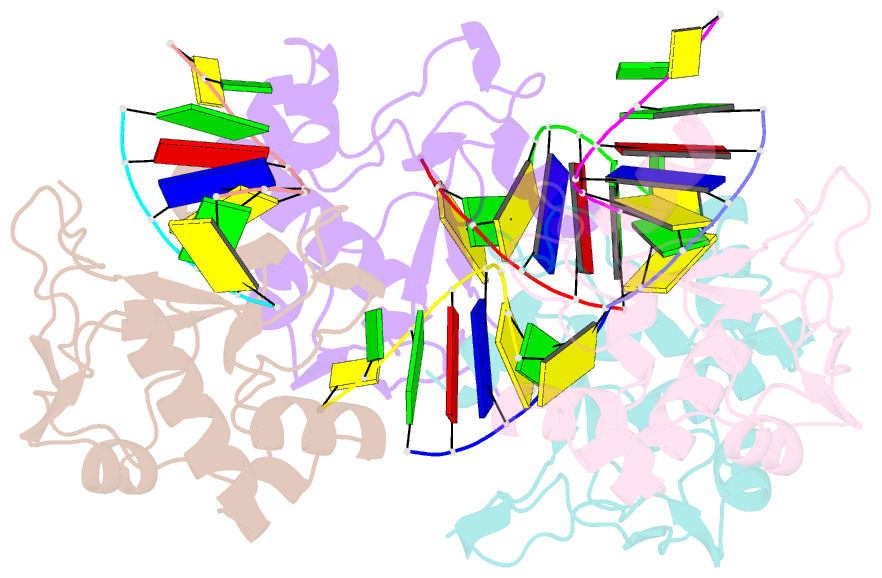
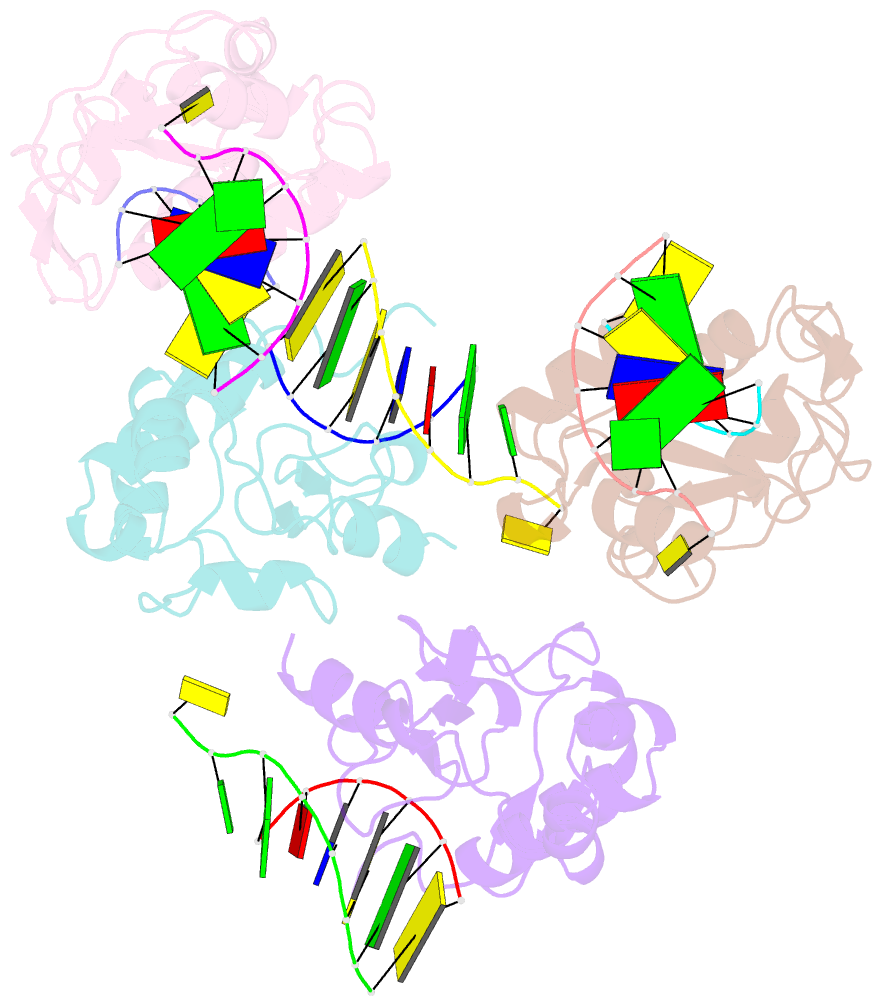
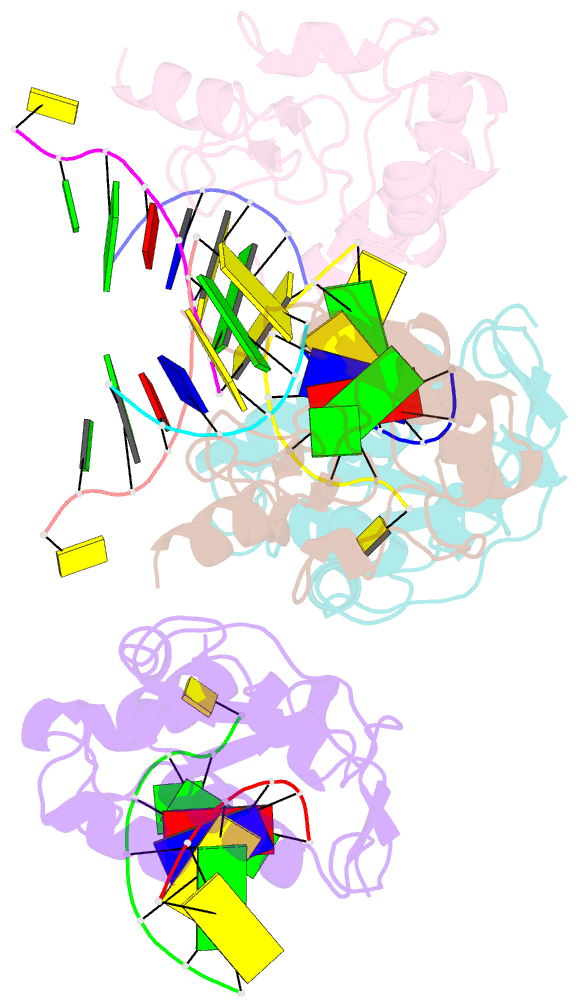
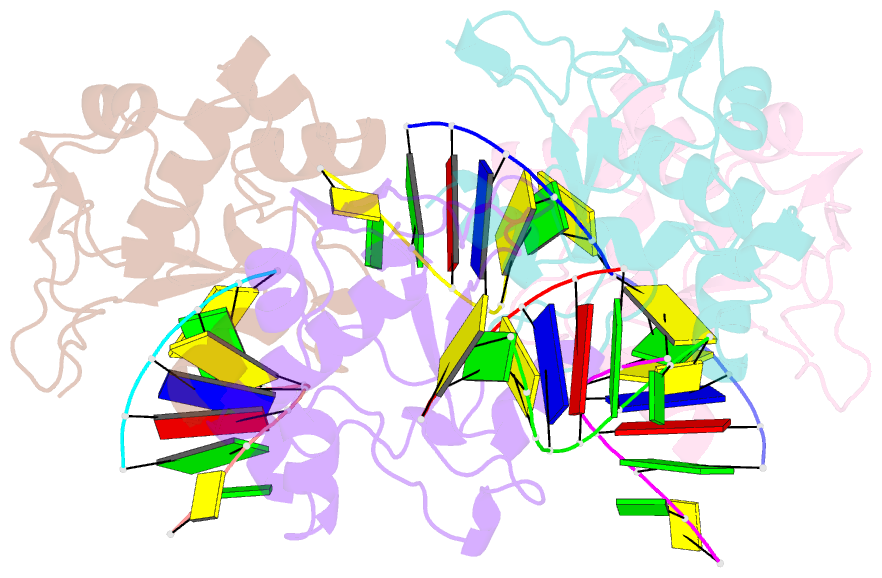
Summary information and primary citation
- PDB-id
- 1v14; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- hydrolase
- Method
- X-ray (2.9 Å)
- Summary
- Crystal structure of the colicin e9, mutant his103ala, in complex with mg+2 and dsDNA (resolution 2.9a)
- Reference
- Mate MJ, Kleanthous C (2004): "Structure-Based Analysis of the Metal-Dependent Mechanism of H-N-H Endonucleases." J.Biol.Chem., 279, 34763. doi: 10.1074/JBC.M403719200.
- Abstract
- Controversy surrounds the metal-dependent mechanism of H-N-H endonucleases, enzymes involved in a variety of biological functions, including intron homing and DNA repair. To address this issue we determined the crystal structures for complexes of the H-N-H motif containing bacterial toxin colicin E9 with Zn(2+), Zn(2+).DNA, and Mg(2+).DNA. The structures show that the rigid V-shaped architecture of the active site does not undergo any major conformational changes on binding to the minor groove of DNA and that the same interactions are made to the nucleic acid regardless of which metal ion is bound to the enzyme. The scissile phosphate contacts the single metal ion of the motif through distortion of the DNA brought about by the insertion of the Arg-96-Glu-100 salt bridge into the minor groove and a network of contacts to the DNA phosphate backbone that straddle the metal site. The Mg(2+)-bound structure reveals an unusual coordination scheme involving two H-N-H histidine residues, His-102 and His-127. The mechanism of DNA cleavage is likely related to that of other single metal ion-dependent endonucleases, such as I-PpoI and Vvn, although in these enzymes the single alkaline earth metal ion is coordinated by oxygen-bearing amino acids. The structures also provide a rationale as to why H-N-H endonucleases are inactive in the presence of Zn(2+) but active with other transition metal ions such as Ni(2+). This is because of coordination of the Zn(2+) ion through a third histidine, His-131. "Active" transition metal ions are those that bind more weakly to the H-N-H motif because of the disengagement of His-131, which we suggest allows a water molecule to complete the catalytic cycle.