Summary information and primary citation
- PDB-id
- 1vtg; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- DNA-antibiotic
- Method
- X-ray (1.67 Å)
- Summary
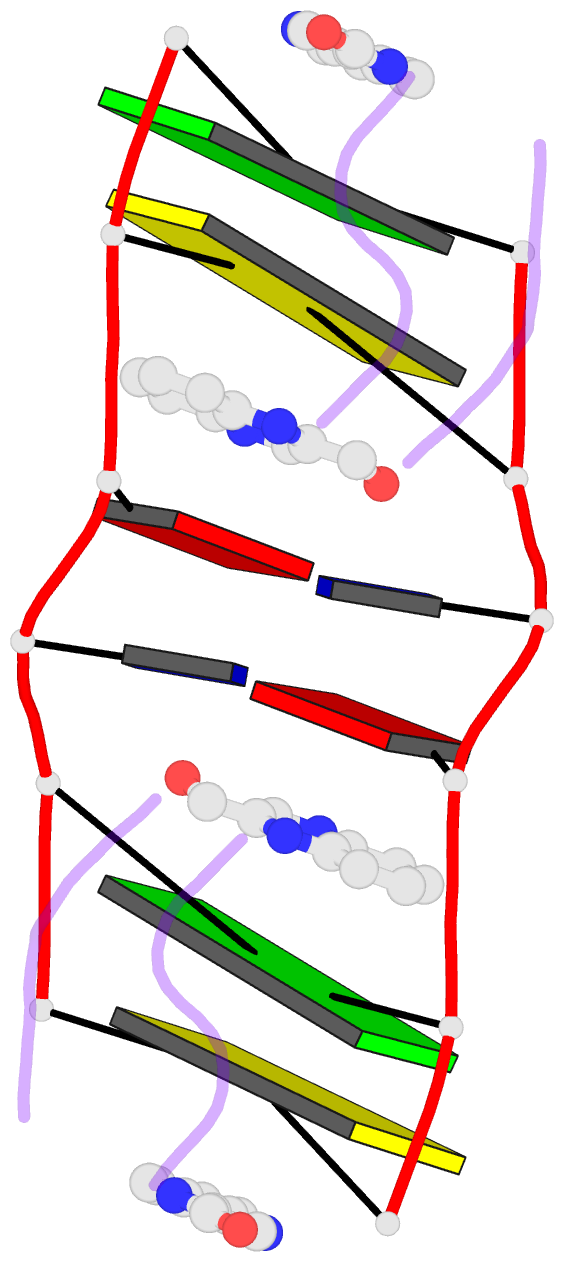
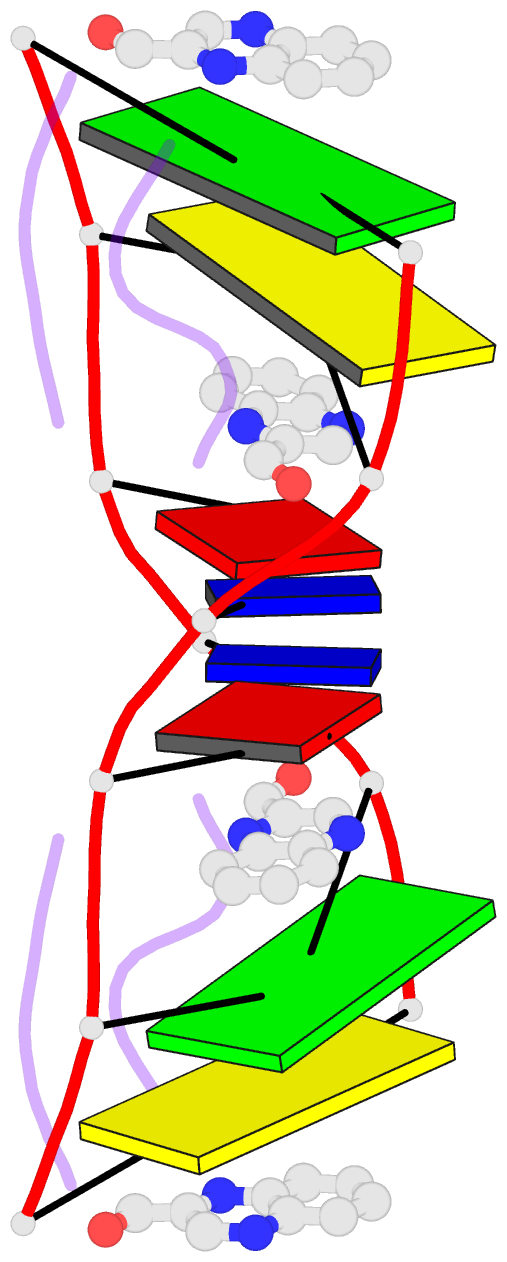
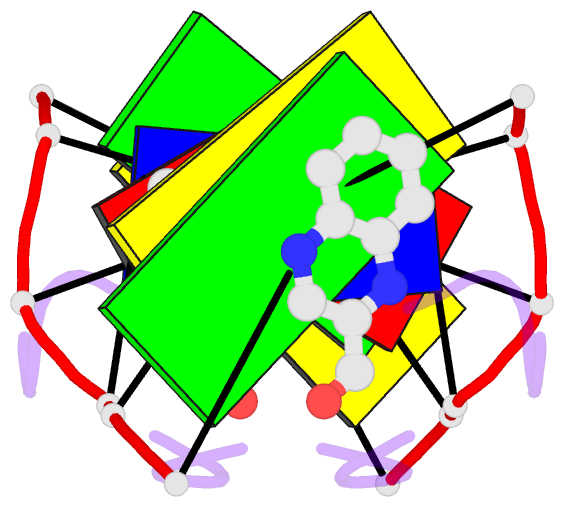
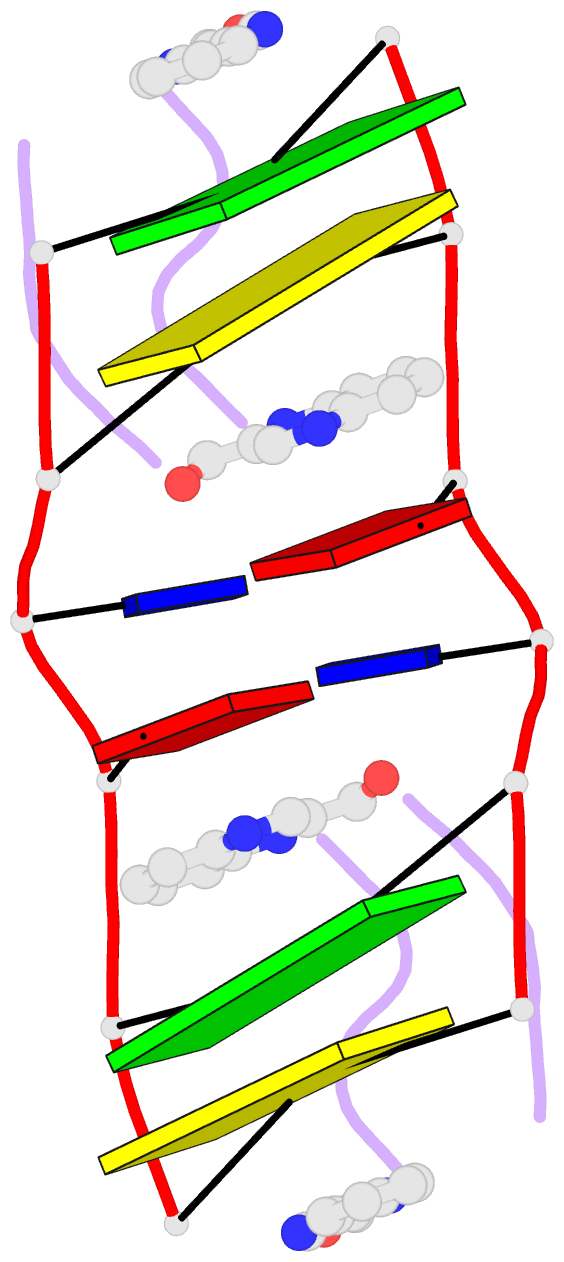
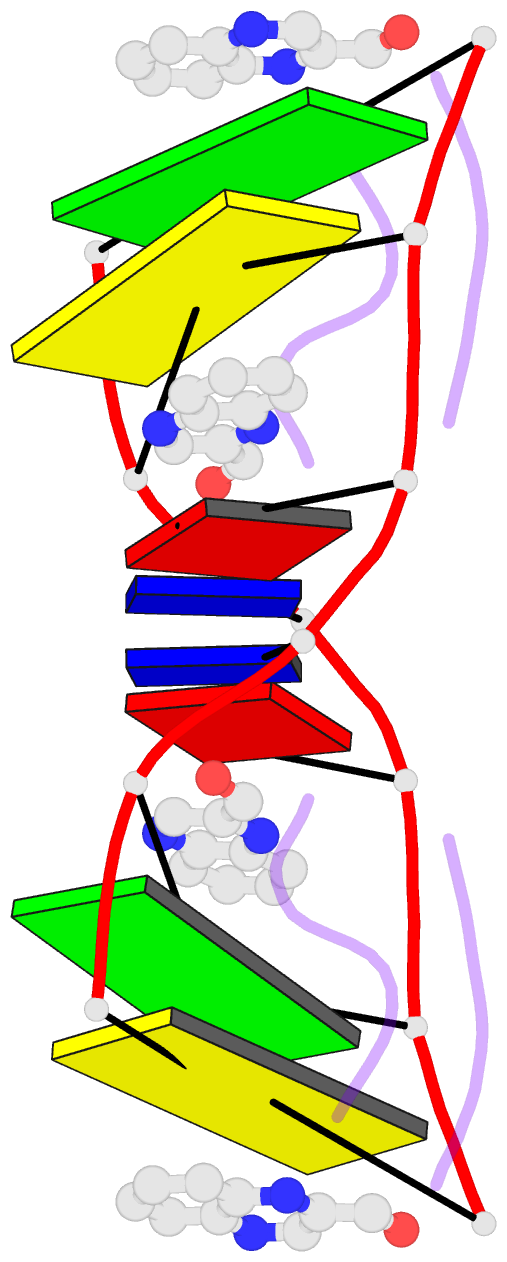
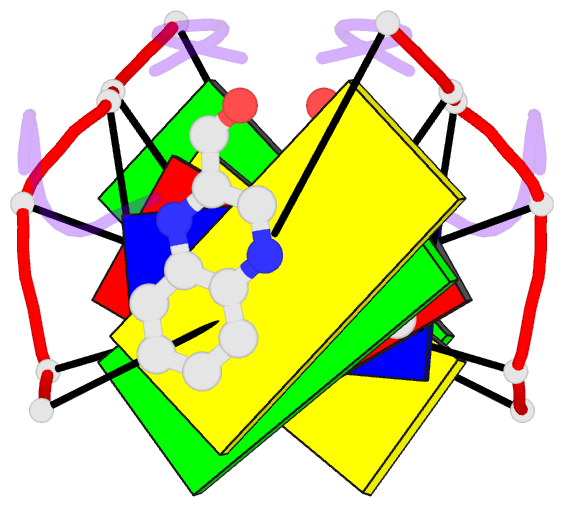
- The molecular structure of a DNA-triostin a complex
- Reference
- Wang AH, Ughetto G, Quigley GJ, Hakoshima T, van der Marel GA, van Boom JH, Rich A (1984): "The molecular structure of a DNA-triostin A complex." Science, 225, 1115-1121.
- Abstract
- The molecular structure of triostin A, a cyclic octadepsipeptide antibiotic, has been solved complexed to a DNA double helical fragment with the sequence CGTACG (C, cytosine; G, guanine; T, thymine; A, adenine). The two planar quinoxaline rings of triostin A bis intercalate on the minor groove of the DNA double helix surrounding the CG base pairs at either end. The alanine residues form hydrogen bonds to the guanines. Base stacking in the DNA is perturbed, and the major binding interaction involves a large number of van der Waals contacts between the peptides and the nucleic acid. The adenine residues in the center are in the syn conformation and are paired to thymine through Hoogsteen base pairing.