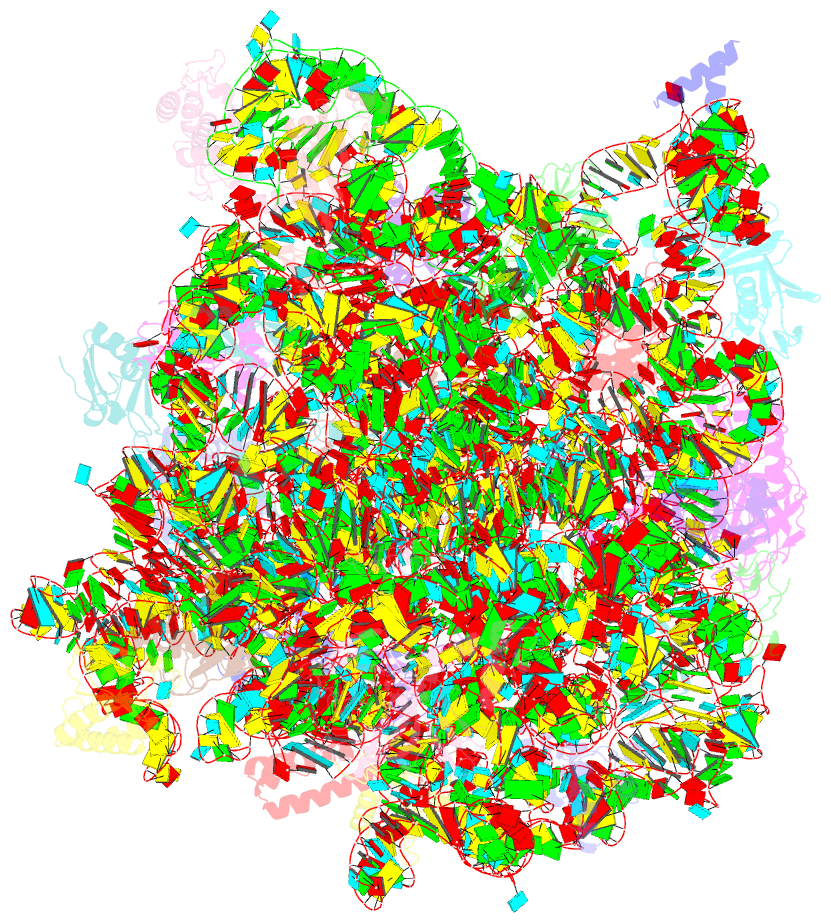
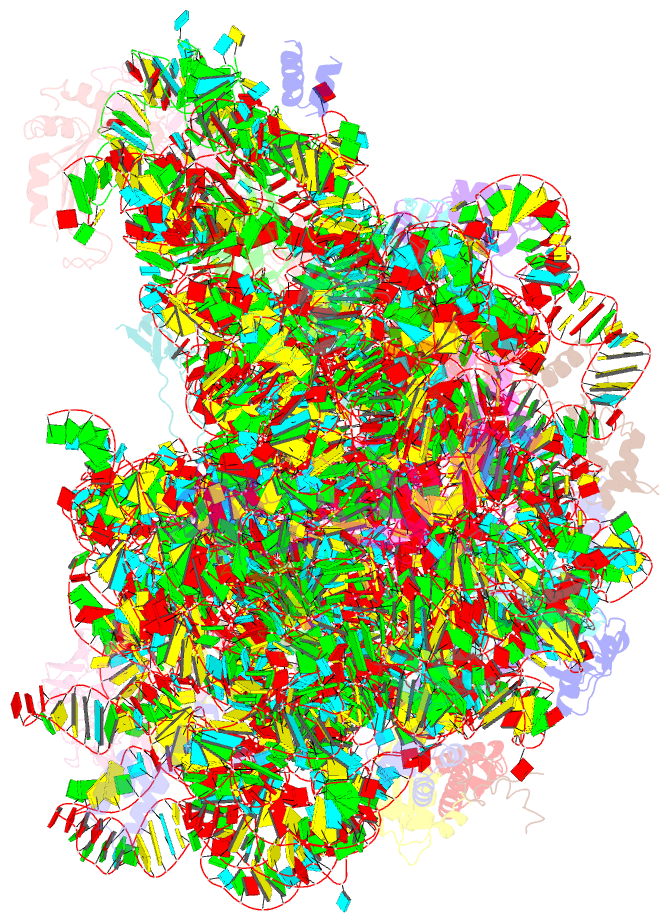
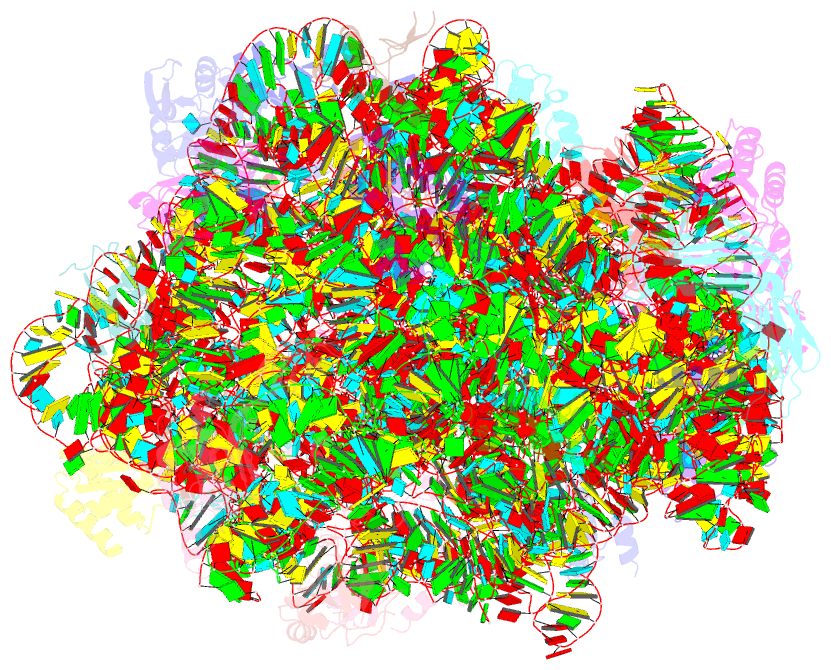
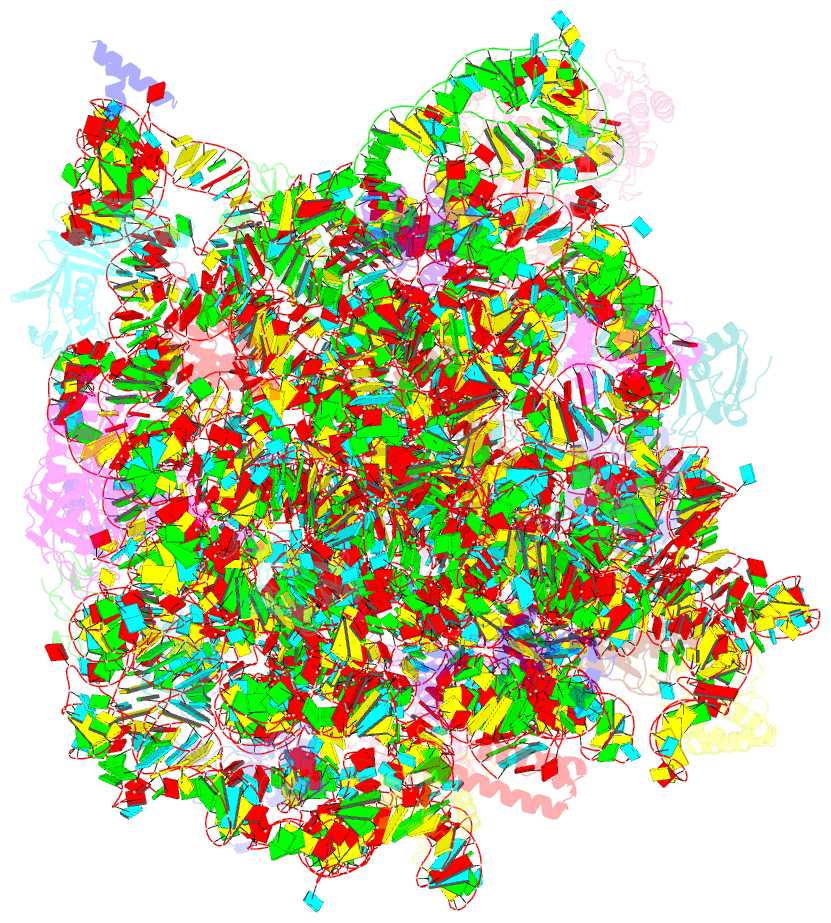
Summary information and primary citation
- PDB-id
- 1w2b; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- ribosome
- Method
- X-ray (3.5 Å)
- Summary
- Trigger factor ribosome binding domain in complex with 50s
- Reference
- Ferbitz L, Maier T, Patzelt H, Bukau B, Deuerling E, Ban N (2004): "Trigger Factor in Complex with the Ribosome Forms a Molecular Cradle for Nascent Proteins." Nature, 431, 590. doi: 10.1038/NATURE02899.
- Abstract
- During protein biosynthesis, nascent polypeptide chains that emerge from the ribosomal exit tunnel encounter ribosome-associated chaperones, which assist their folding to the native state. Here we present a 2.7 A crystal structure of Escherichia coli trigger factor, the best-characterized chaperone of this type, together with the structure of its ribosome-binding domain in complex with the Haloarcula marismortui large ribosomal subunit. Trigger factor adopts a unique conformation resembling a crouching dragon with separated domains forming the amino-terminal ribosome-binding 'tail', the peptidyl-prolyl isomerase 'head', the carboxy-terminal 'arms' and connecting regions building up the 'back'. From its attachment point on the ribosome, trigger factor projects the extended domains over the exit of the ribosomal tunnel, creating a protected folding space where nascent polypeptides may be shielded from proteases and aggregation. This study sheds new light on our understanding of co-translational protein folding, and suggests an unexpected mechanism of action for ribosome-associated chaperones.