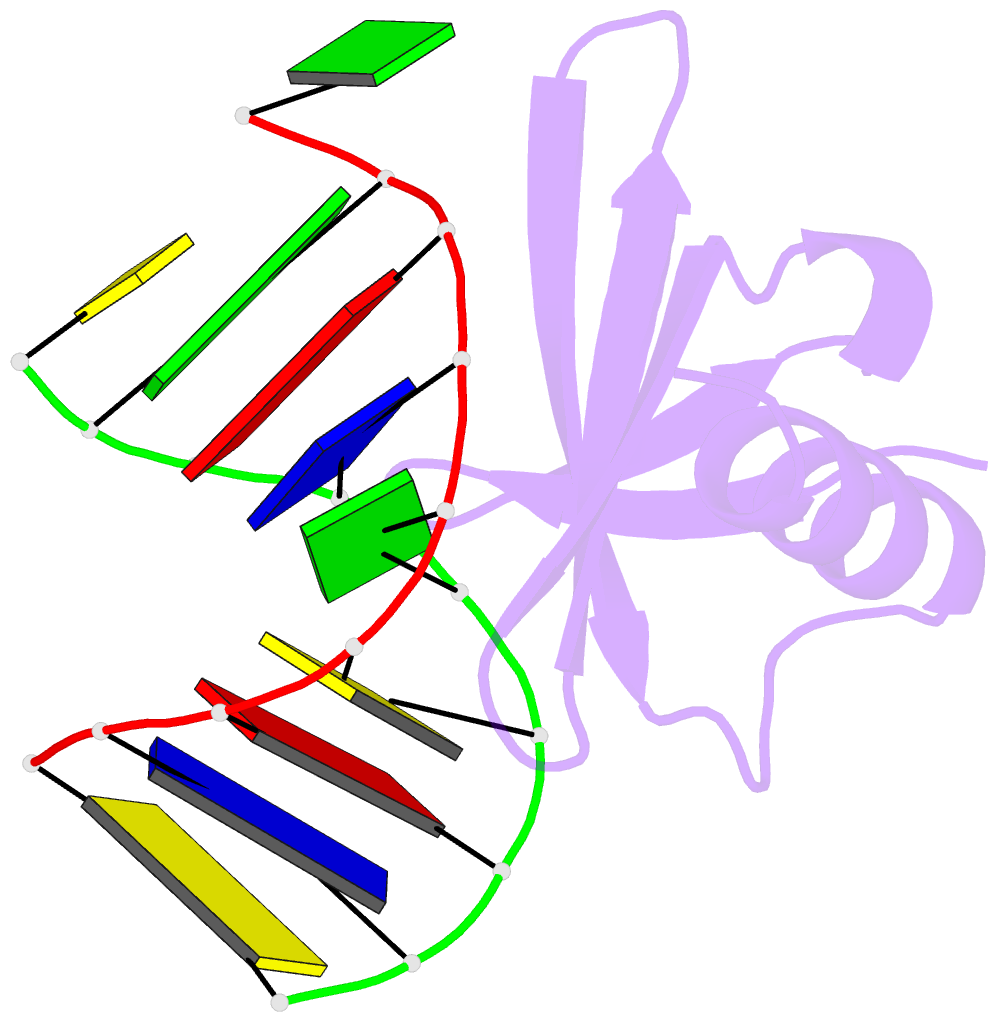
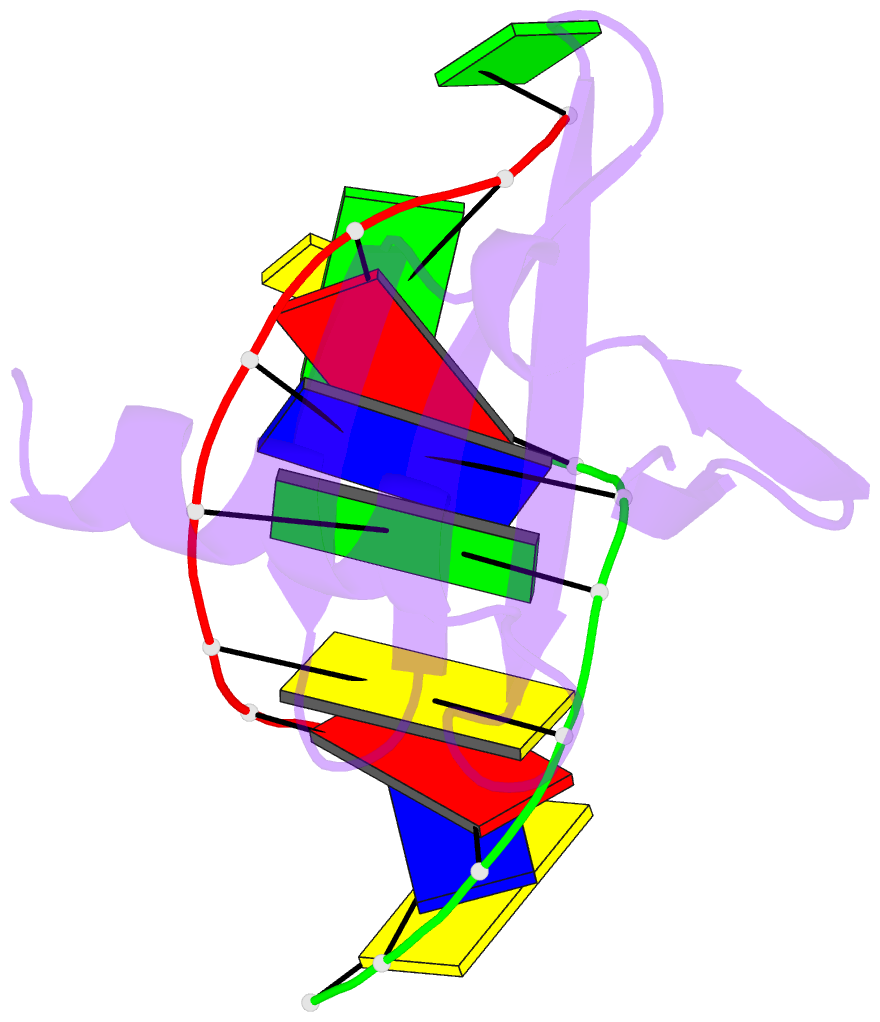
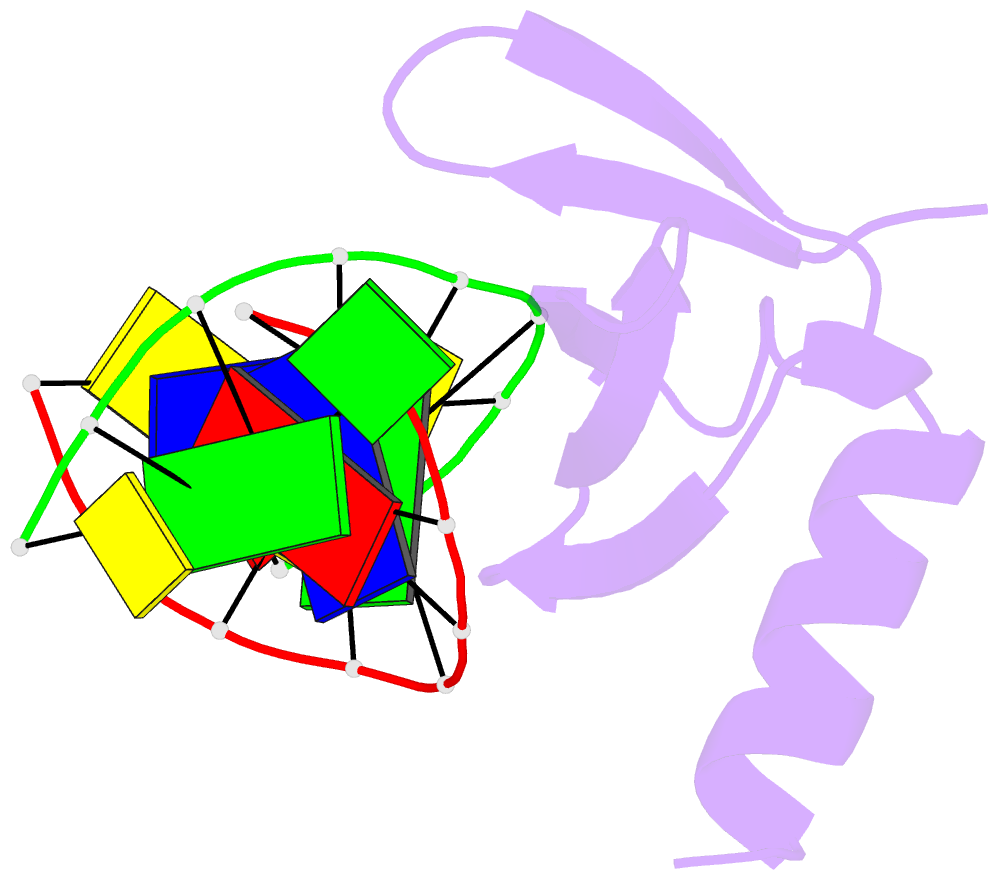
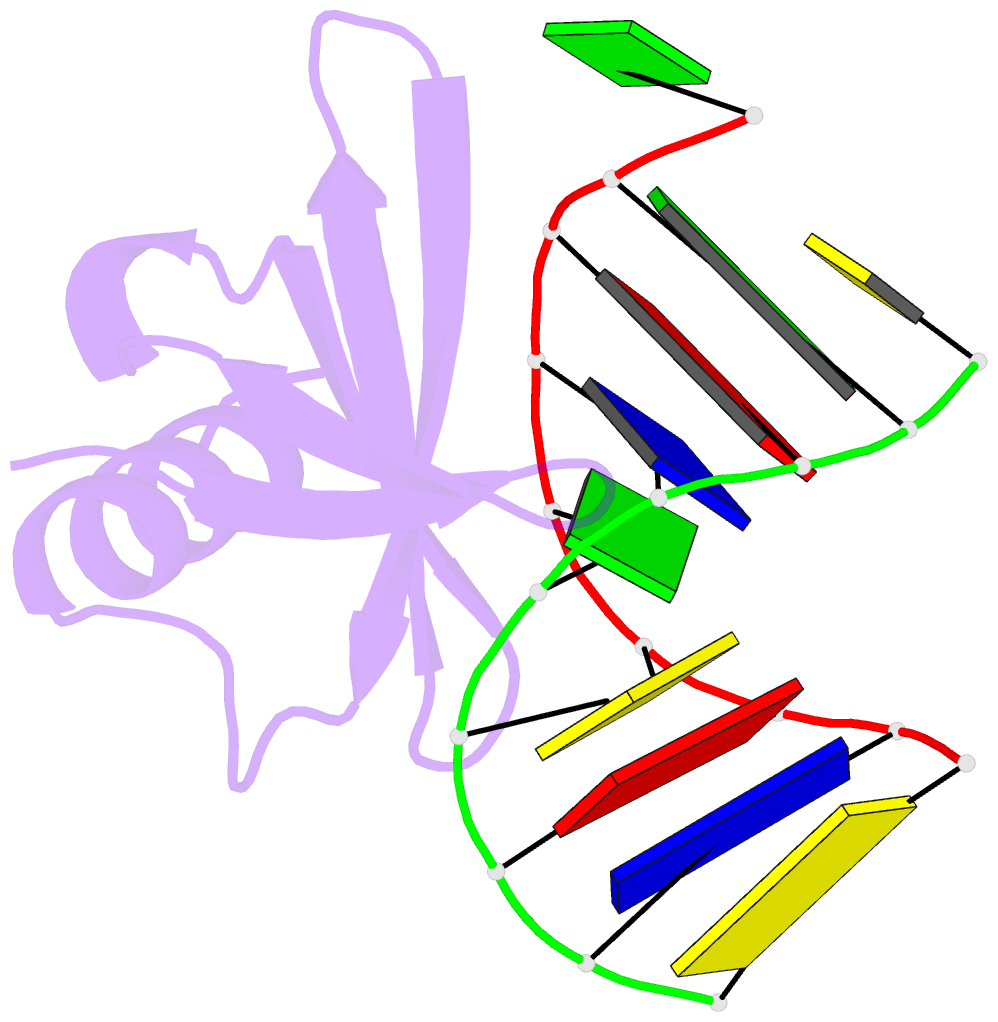
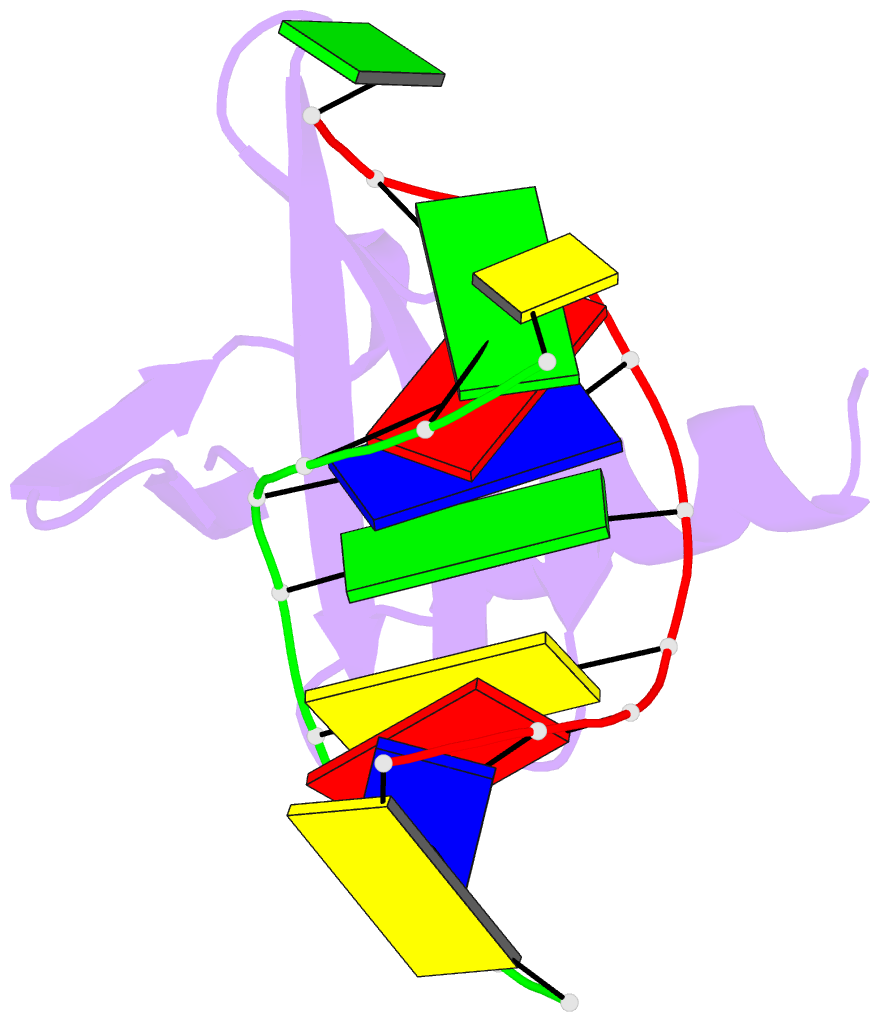
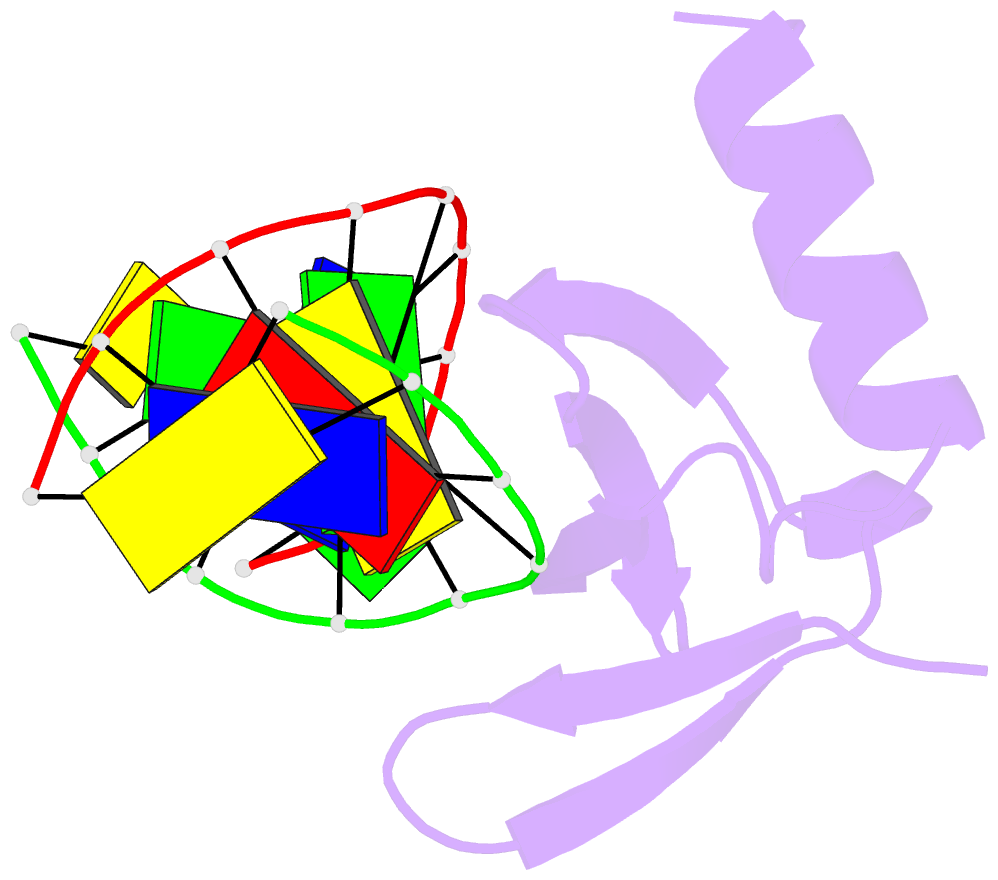
Summary information and primary citation
- PDB-id
- 1wd1; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- structural protein-DNA
- Method
- X-ray (2.2 Å)
- Summary
- Crystal structures of the hyperthermophilic chromosomal protein sac7d in complex with DNA decamers
- Reference
- Ko TP, Chu HM, Chen CY, Chou CC, Wang AH (2004): "Structures of the hyperthermophilic chromosomal protein Sac7d in complex with DNA decamers." Acta Crystallogr.,Sect.D, 60, 1381-1387. doi: 10.1107/S0907444904012065.
- Abstract
- The protein Sac7d belongs to a class of small chromosomal proteins from the hyperthermophilic archaeon Sulfolobus acidocaldarius. Two new crystal forms of Sac7d in complex with the DNA decamers CCTATATAGG and CCTACGTAGG were obtained and their structures were determined by molecular replacement. The refined models yielded R/Rfree values of 0.221/0.257 and 0.248/0.290 at 1.9 and 2.2 A resolution, respectively. The protein structures are similar to the previously determined structure of Sac7d-GCGATCGC (PDB code 1azp), but the DNA molecules are more bent overall, by 14-20 degrees. The relative positions of the Sac7d protein and the bound DNA also differ by rotations of 6-10 degrees and translations of 1.0-2.4 A. In addition to the water molecules in the central cavity, three additional conserved water molecules are found that mediate the protein-DNA interactions. The decamer DNA fragments form virtual double helices in the crystal, with a unit length of eight base pairs. The molecular packing of the new crystal forms differs from that of 1azp. The terminal nucleotides are opened up and form triple base pairs with other DNA molecules. Through lattice contacts, the Sac7d molecule also makes additional interactions with DNA, whereas only limited protein-protein interactions are seen.