Summary information and primary citation
- PDB-id
- 1wet; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transcription-DNA
- Method
- X-ray (2.6 Å)
- Summary
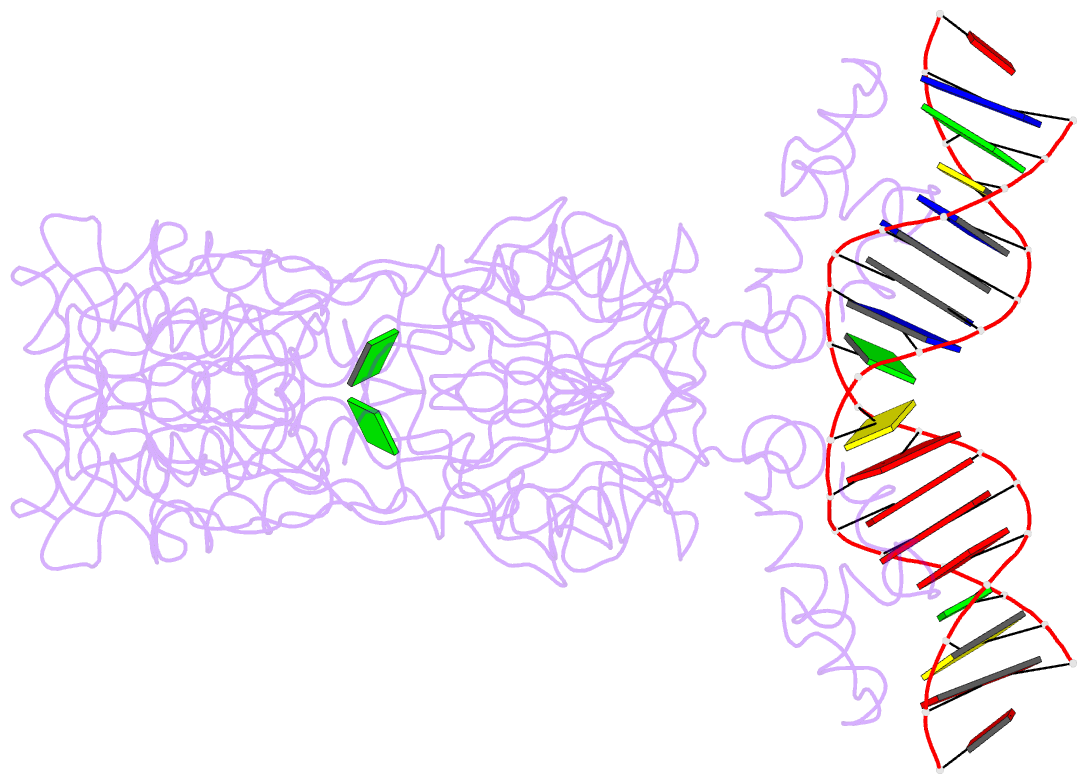
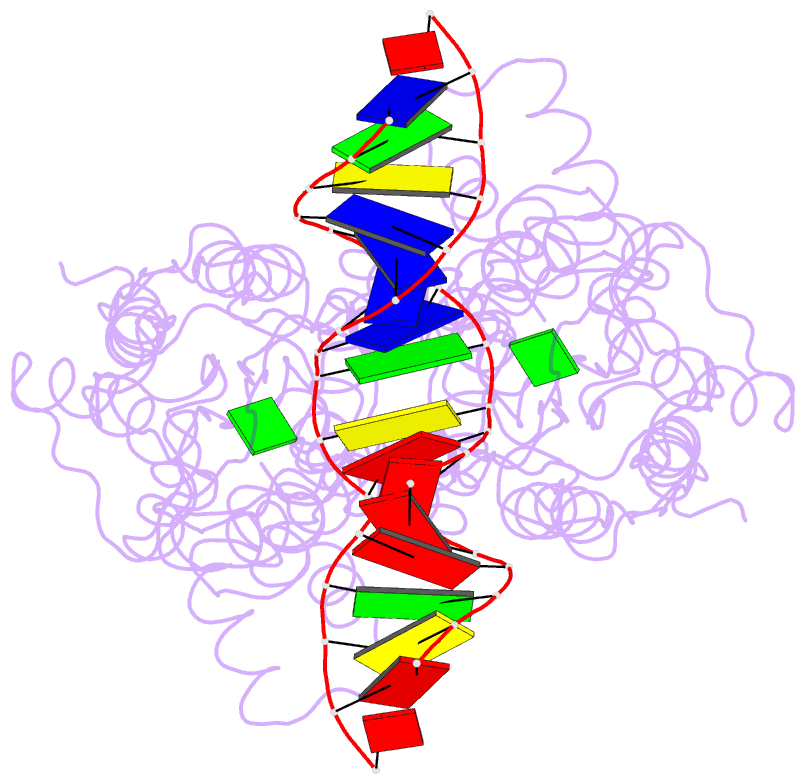
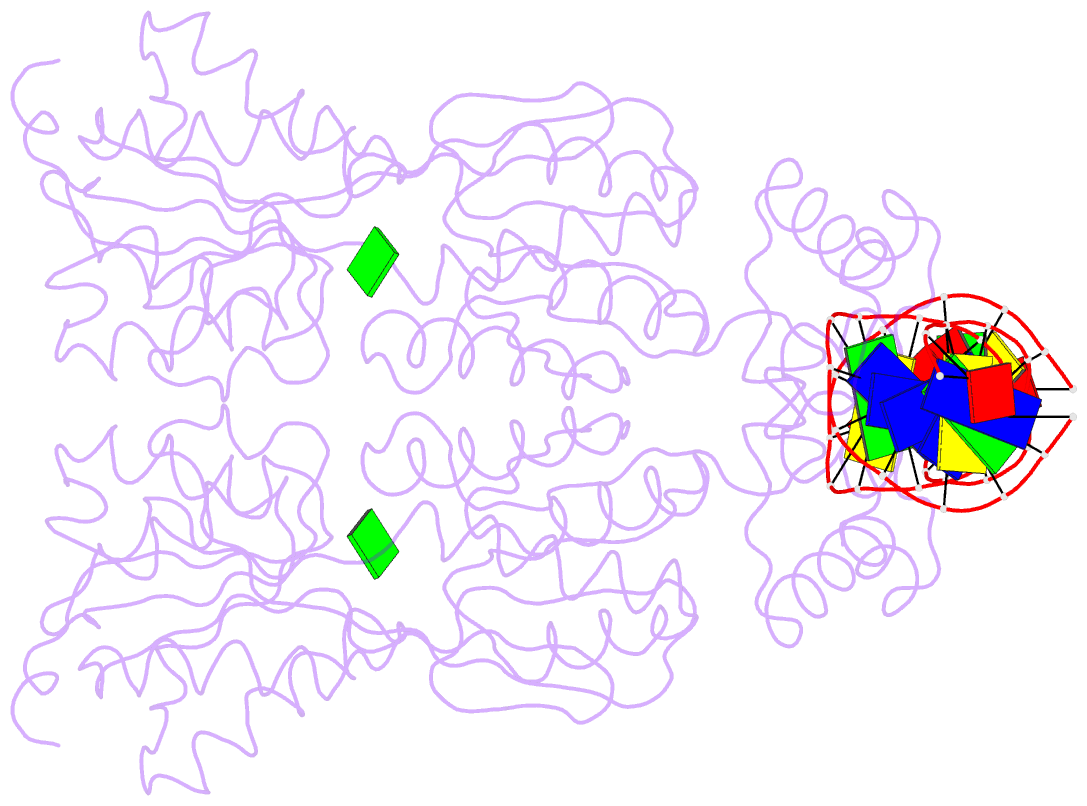
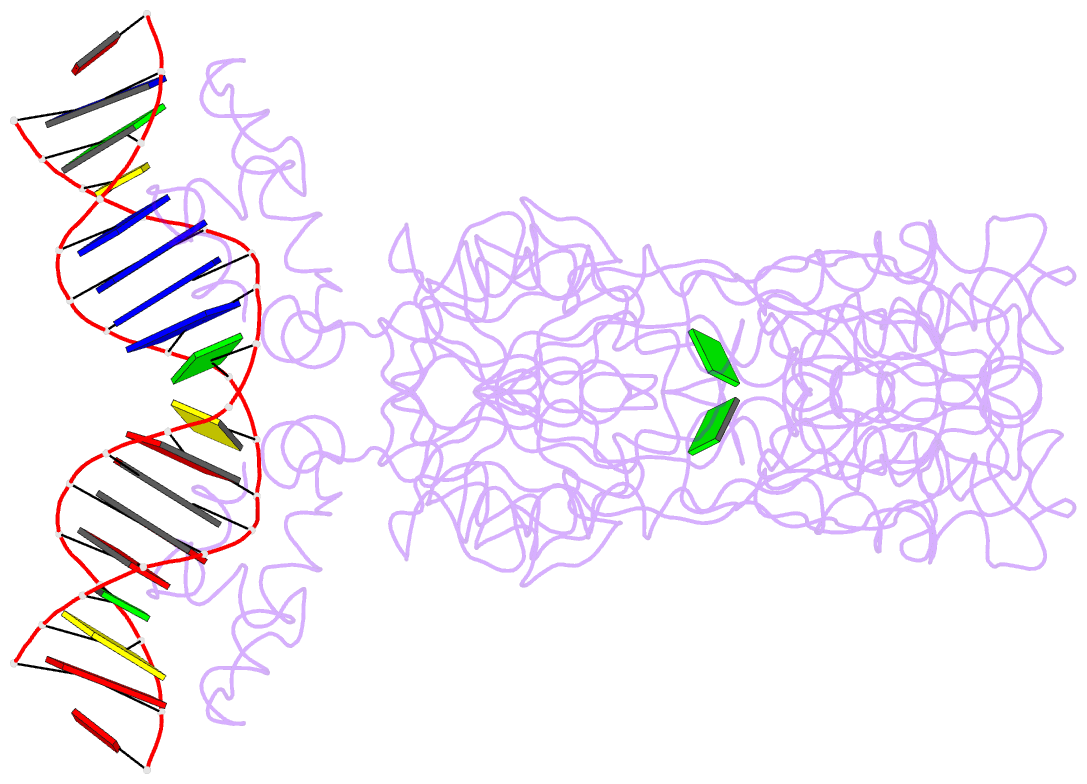
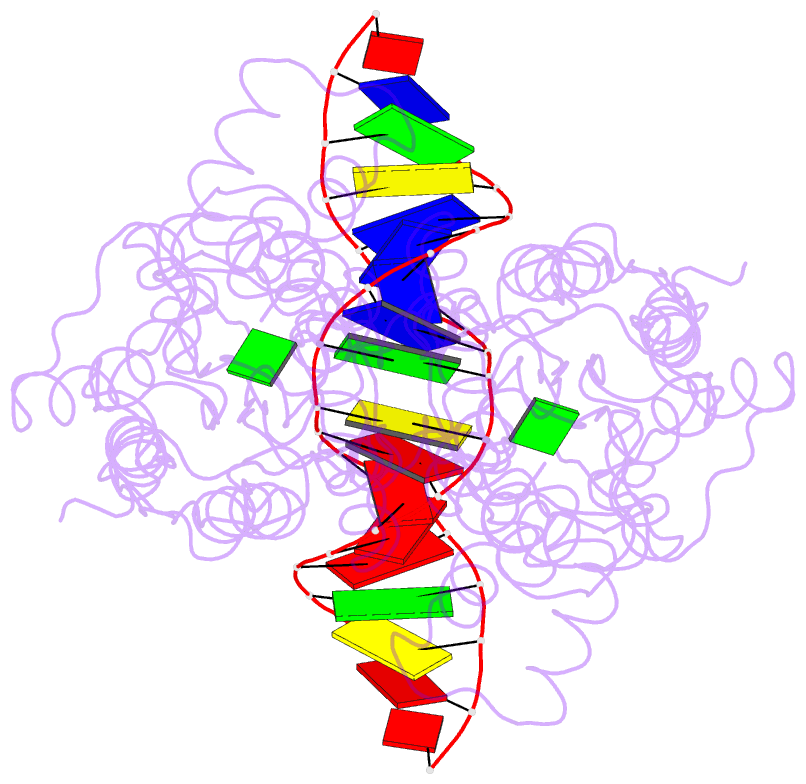
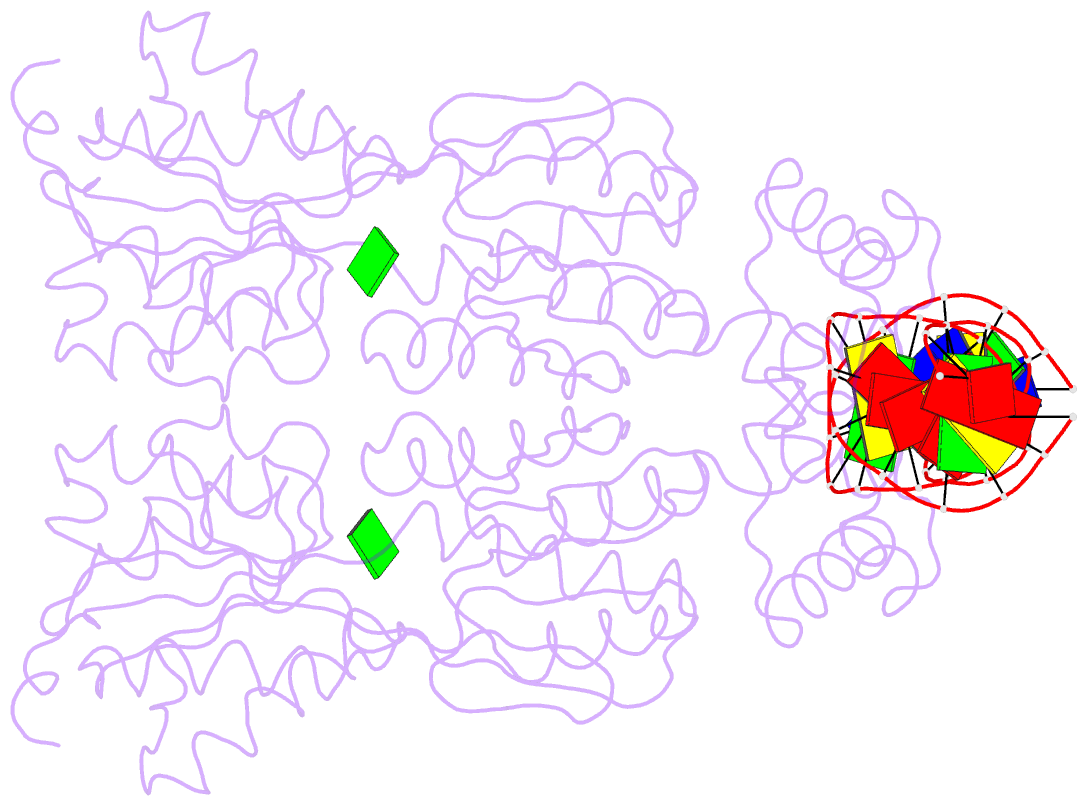
- Structure of the purr-guanine-purf operator complex
- Reference
- Schumacher MA, Glasfeld A, Zalkin H, Brennan RG (1997): "The X-ray structure of the PurR-guanine-purF operator complex reveals the contributions of complementary electrostatic surfaces and a water-mediated hydrogen bond to corepressor specificity and binding affinity." J.Biol.Chem., 272, 22648-22653. doi: 10.1074/jbc.272.36.22648.
- Abstract
- The purine repressor, PurR, is the master regulatory protein of de novo purine nucleotide biosynthesis in Escherichia coli. This dimeric transcription factor is activated to bind to cognate DNA operator sites by initially binding either of its physiologically relevant, high affinity corepressors, hypoxanthine (Kd = 9.3 microM) or guanine (Kd = 1.5 microM). Here, we report the 2.5-A crystal structure of the PurR-guanine-purF operator ternary complex and complete the atomic description of 6-oxopurine-induced repression by PurR. As anticipated, the structure of the PurR-guanine-purF operator complex is isomorphous to the PurR-hypoxanthine-purF operator complex, and their protein-DNA and protein-corepressor interactions are nearly identical. The former finding confirms the use of an identical allosteric DNA-binding mechanism whereby corepressor binding 40 A from the DNA-binding domain juxtaposes the hinge regions of each monomer, thus favoring the formation and insertion of the critical minor groove-binding hinge helices. Strikingly, the higher binding affinity of guanine for PurR and the ability of PurR to discriminate against 2-oxopurines do not result from direct protein-ligand interactions, but rather from a water-mediated contact with the exocyclic N-2 of guanine, which dictates the presence of a donor group on the corepressor, and the better electrostatic complementarity of the guanine base and the corepressor-binding pocket.