Summary information and primary citation
- PDB-id
- 1x9n; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- ligase-DNA
- Method
- X-ray (3.0 Å)
- Summary
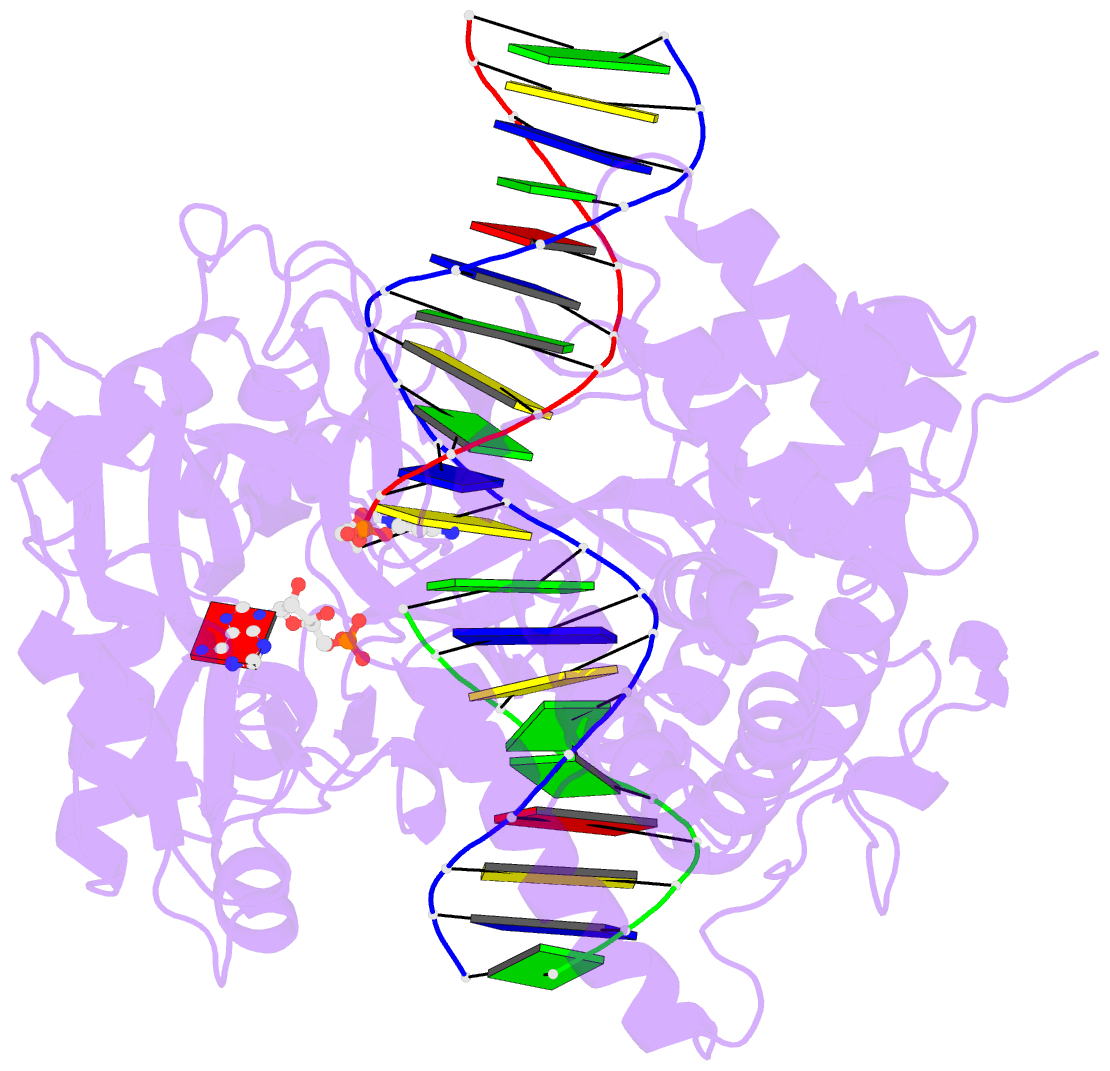
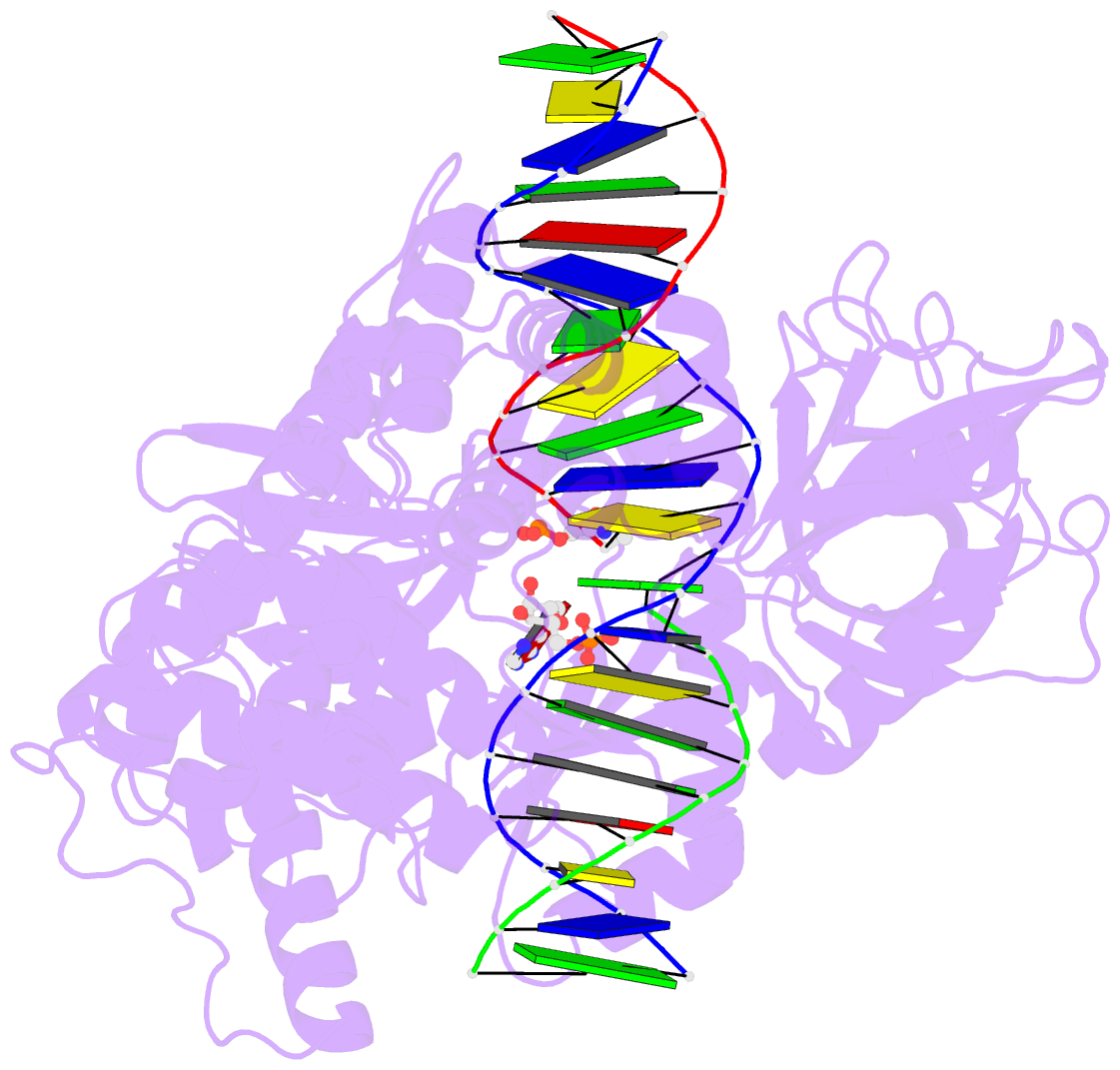
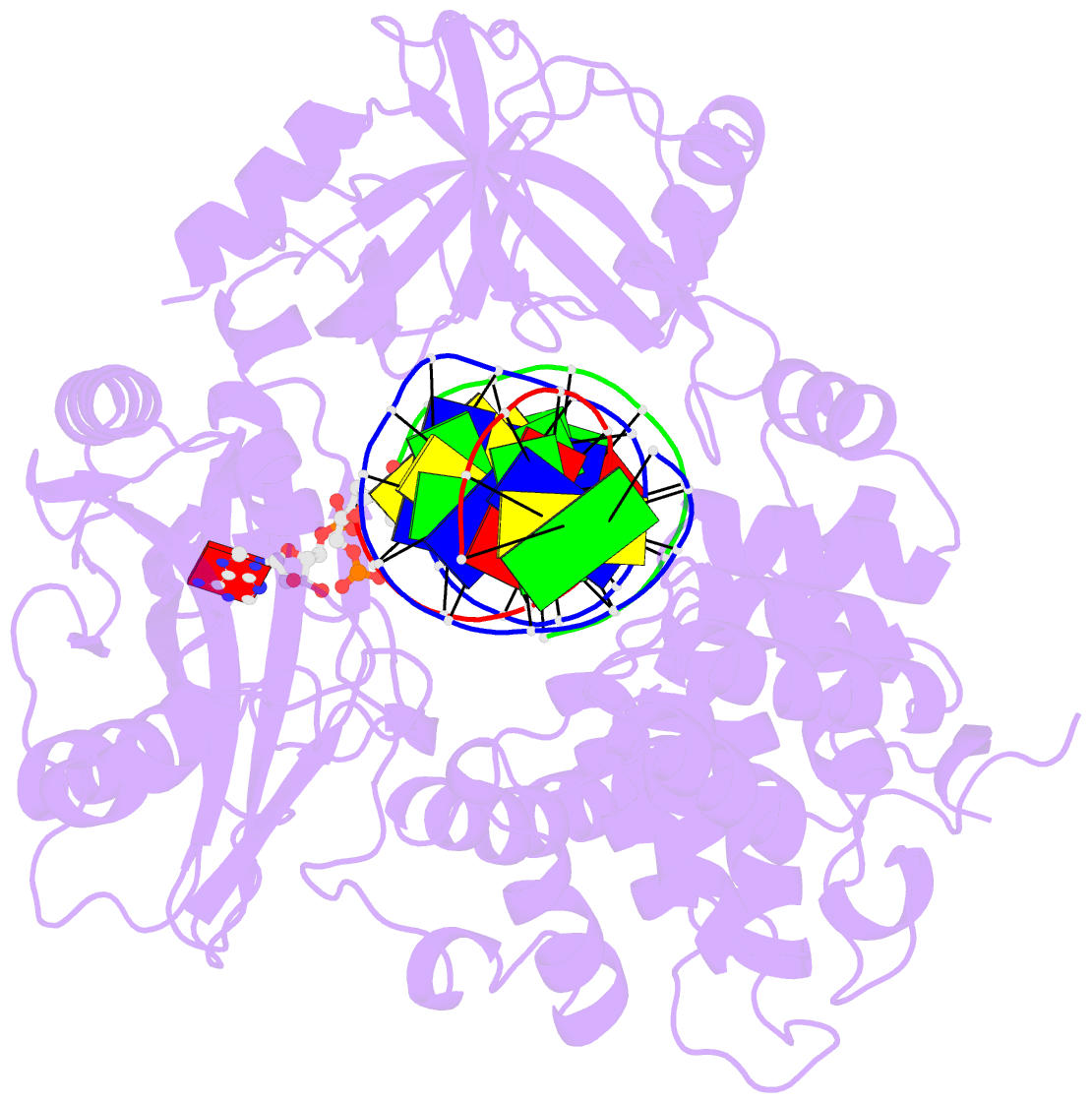
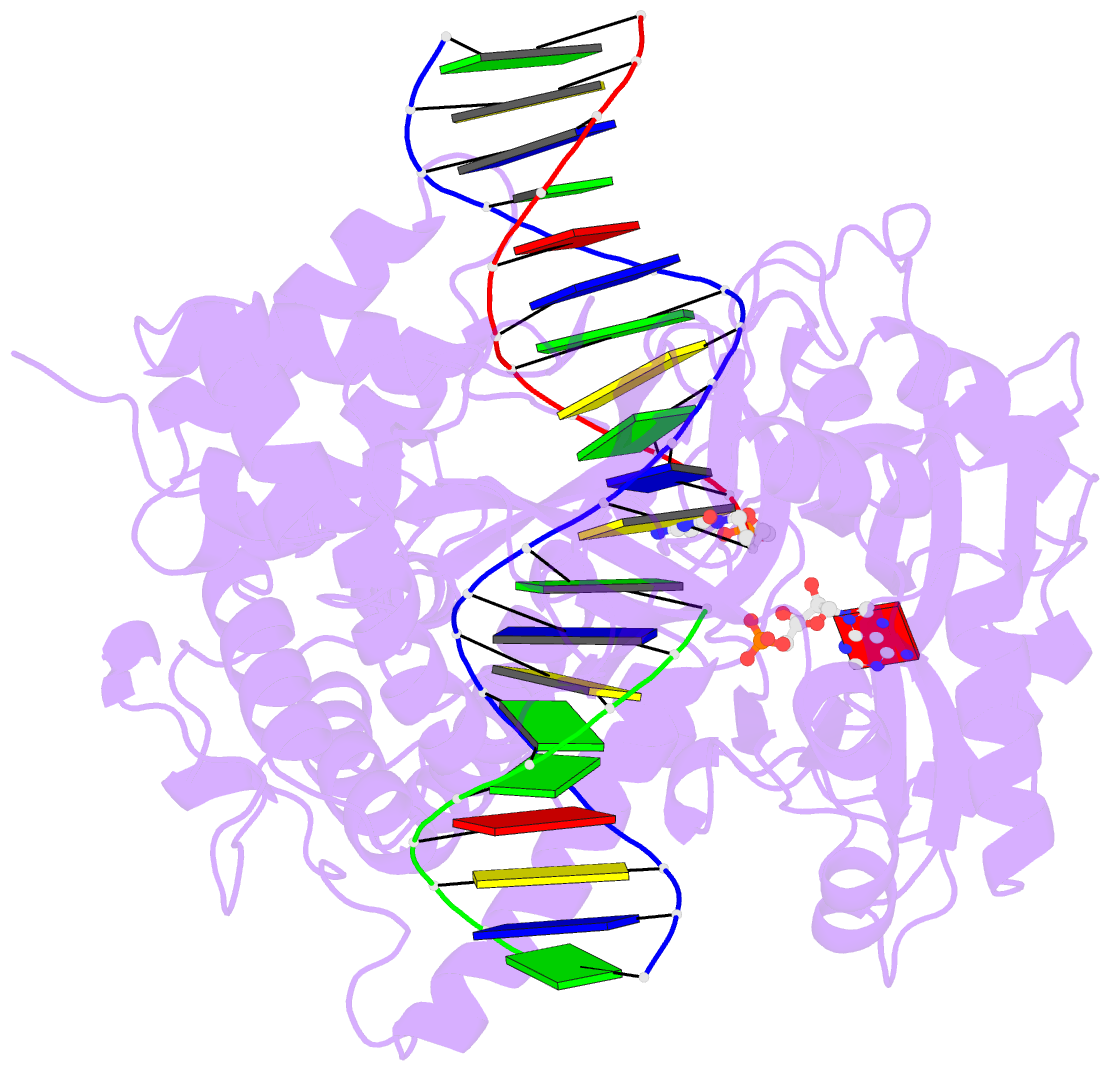
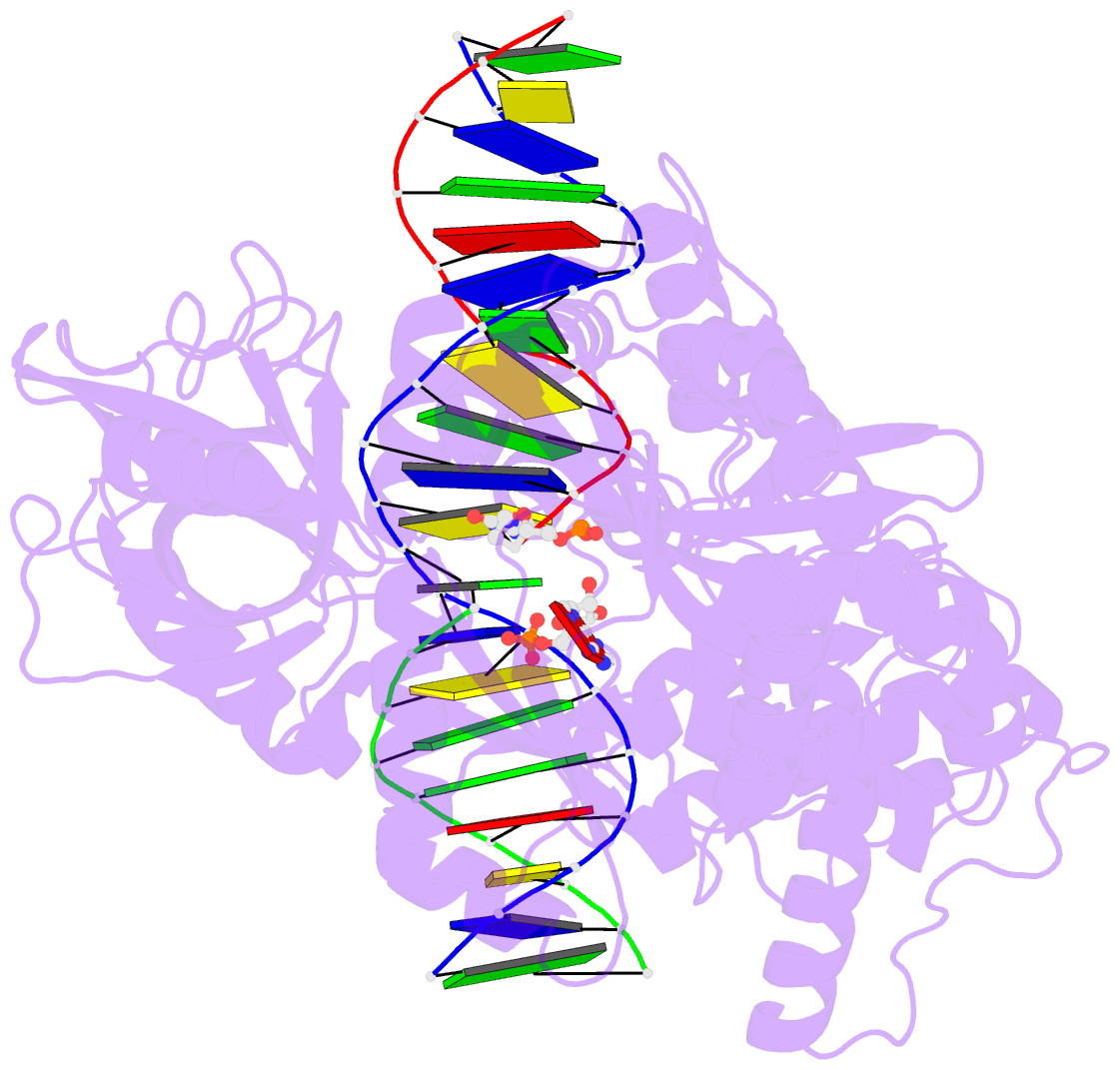
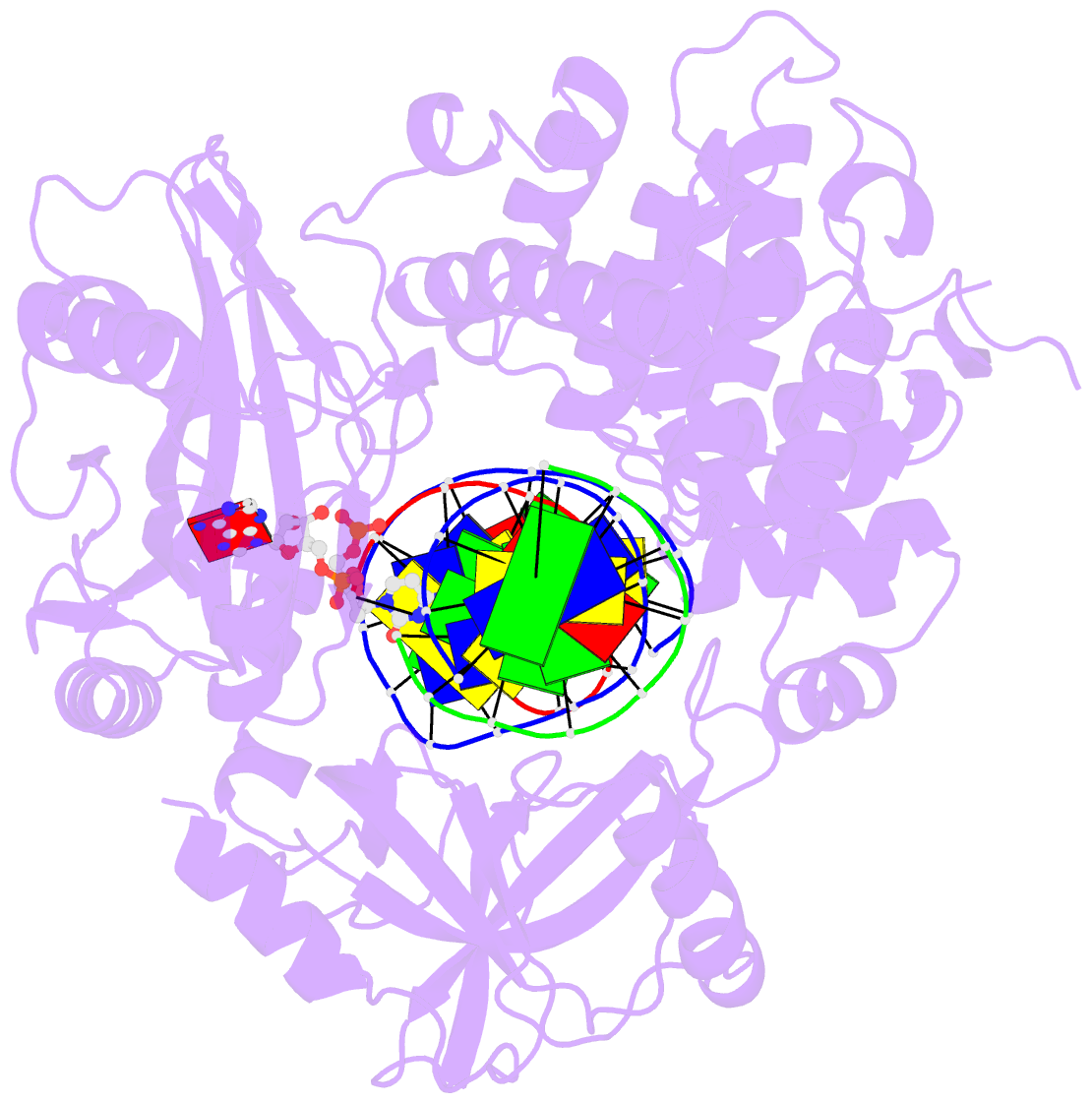
- Crystal structure of human DNA ligase i bound to 5'-adenylated, nicked DNA
- Reference
- Pascal JM, O'Brien PJ, Tomkinson AE, Ellenberger T (2004): "Human DNA ligase I completely encircles and partially unwinds nicked DNA." Nature, 432, 473-478. doi: 10.1038/nature03082.
- Abstract
- The end-joining reaction catalysed by DNA ligases is required by all organisms and serves as the ultimate step of DNA replication, repair and recombination processes. One of three well characterized mammalian DNA ligases, DNA ligase I, joins Okazaki fragments during DNA replication. Here we report the crystal structure of human DNA ligase I (residues 233 to 919) in complex with a nicked, 5' adenylated DNA intermediate. The structure shows that the enzyme redirects the path of the double helix to expose the nick termini for the strand-joining reaction. It also reveals a unique feature of mammalian ligases: a DNA-binding domain that allows ligase I to encircle its DNA substrate, stabilizes the DNA in a distorted structure, and positions the catalytic core on the nick. Similarities in the toroidal shape and dimensions of DNA ligase I and the proliferating cell nuclear antigen sliding clamp are suggestive of an extensive protein-protein interface that may coordinate the joining of Okazaki fragments.