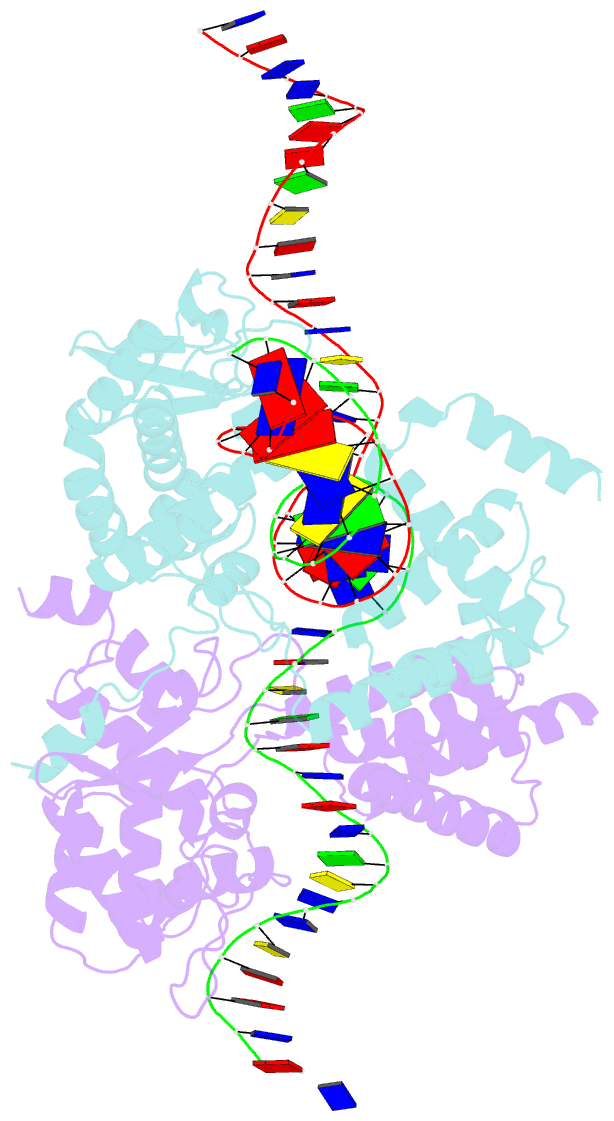
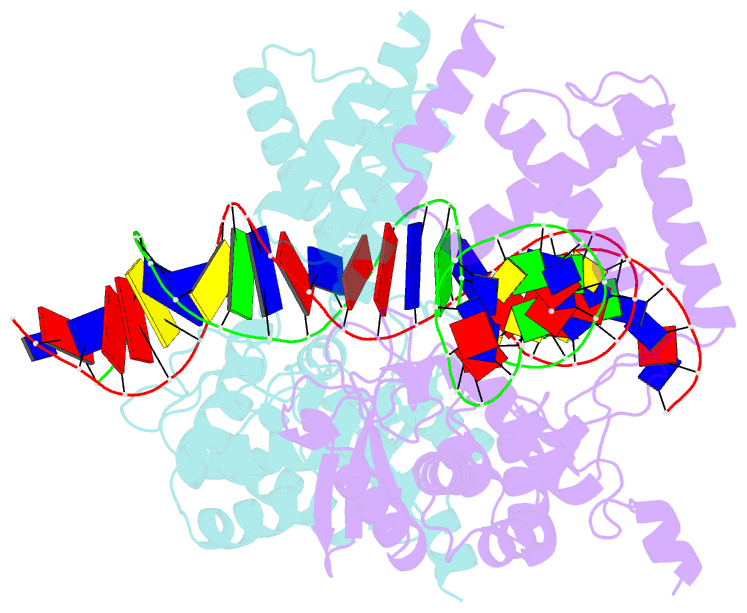
Summary information and primary citation
- PDB-id
- 1xo0; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- hydrolase, ligase-DNA
- Method
- X-ray (2.0 Å)
- Summary
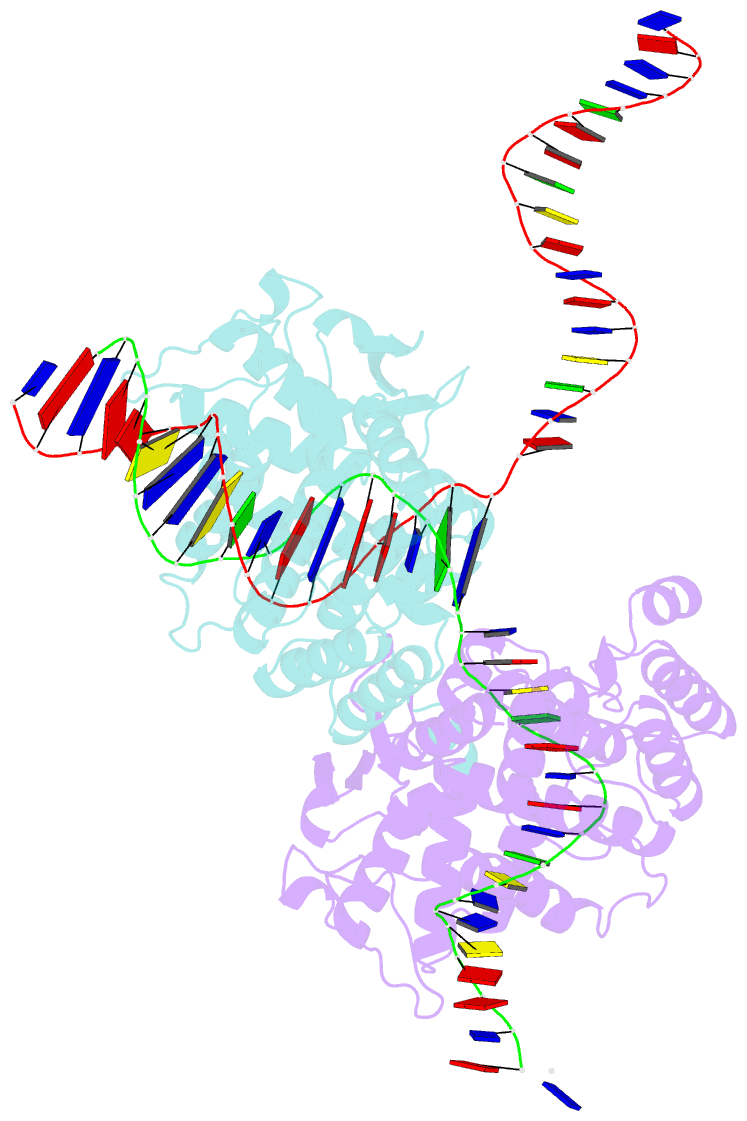
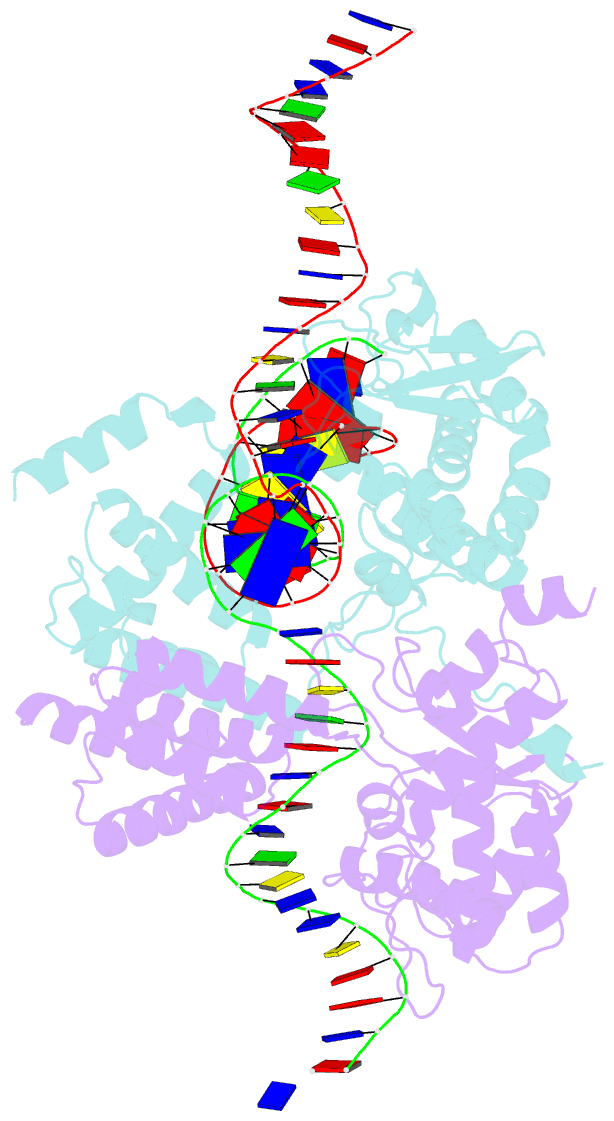
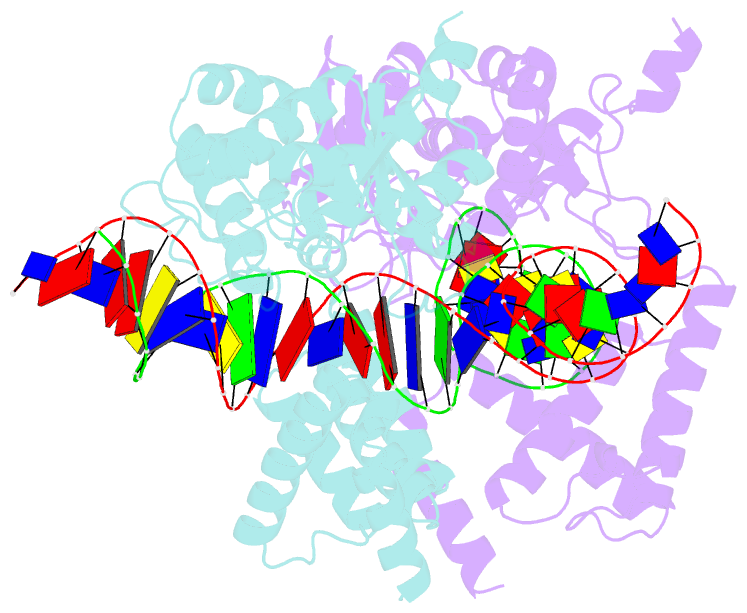
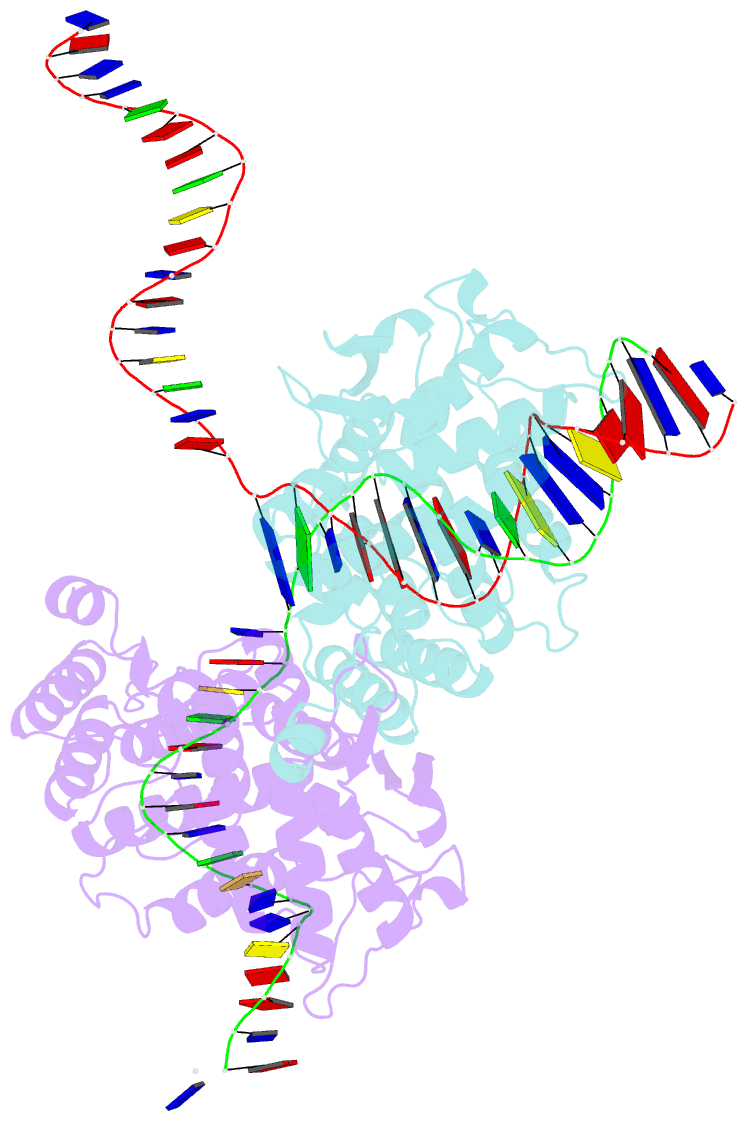
- High resolution structure of the holliday junction intermediate in cre-loxp site-specific recombination
- Reference
- Ghosh K, Lau CK, Guo F, Segall AM, Van Duyne GD (2005): "Peptide trapping of the Holliday junction intermediate in Cre-loxP site-specific recombination." J.Biol.Chem., 280, 8290-8299. doi: 10.1074/jbc.M411668200.
- Abstract
- Cre recombinase is a prototypical member of the tyrosine recombinase family of site-specific recombinases. Members of this family of enzymes catalyze recombination between specific DNA sequences by cleaving and exchanging one pair of strands between the two substrate sites to form a 4-way Holliday junction (HJ) intermediate and then resolve the HJ intermediate to recombinant products by a second round of strand exchanges. Recently, hexapeptide inhibitors have been described that are capable of blocking the second strand exchange step in the tyrosine recombinase recombination pathway, leading to an accumulation of the HJ intermediate. These peptides are active in the lambda-integrase, Cre recombinase, and Flp recombinase systems and are potentially important tools for both in vitro mechanistic studies and as in vivo probes of cellular function. Here we present biochemical and crystallographic data that support a model where the peptide inhibitor binds in the center of the recombinase-bound DNA junction and interacts with solvent-exposed bases near the junction branch point. Peptide binding induces large conformational changes in the DNA strands of the HJ intermediate, which affect the active site geometries in the recombinase subunits.