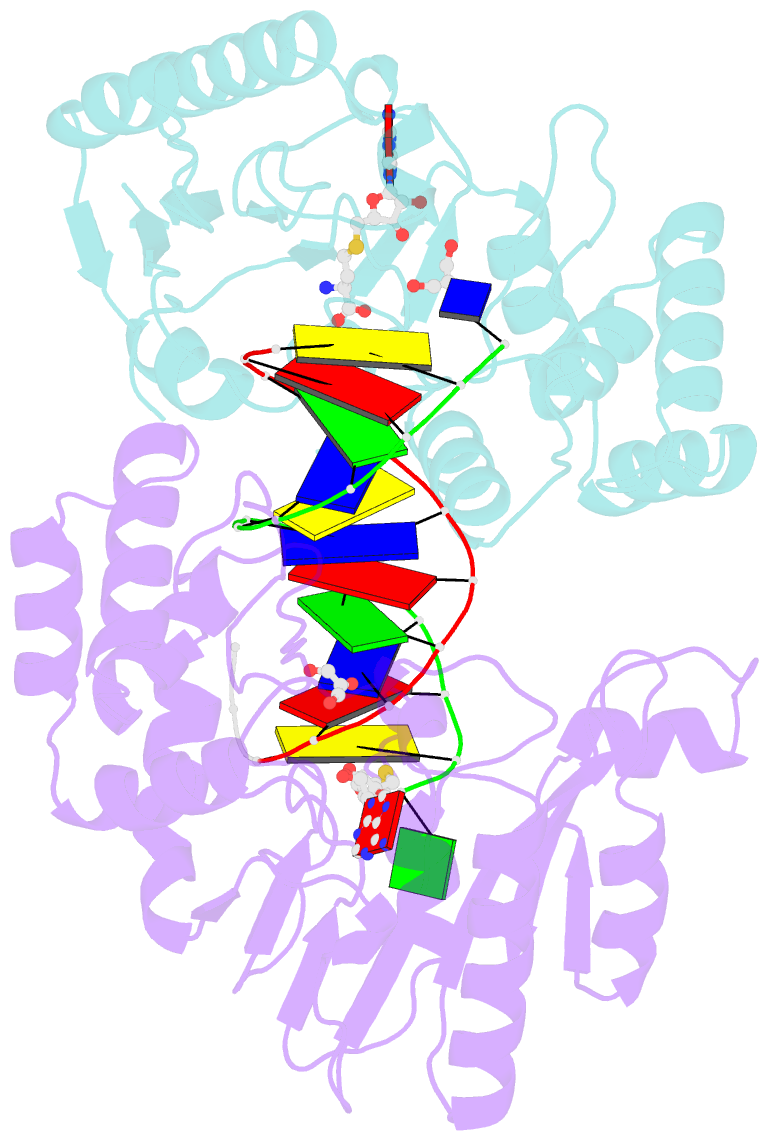
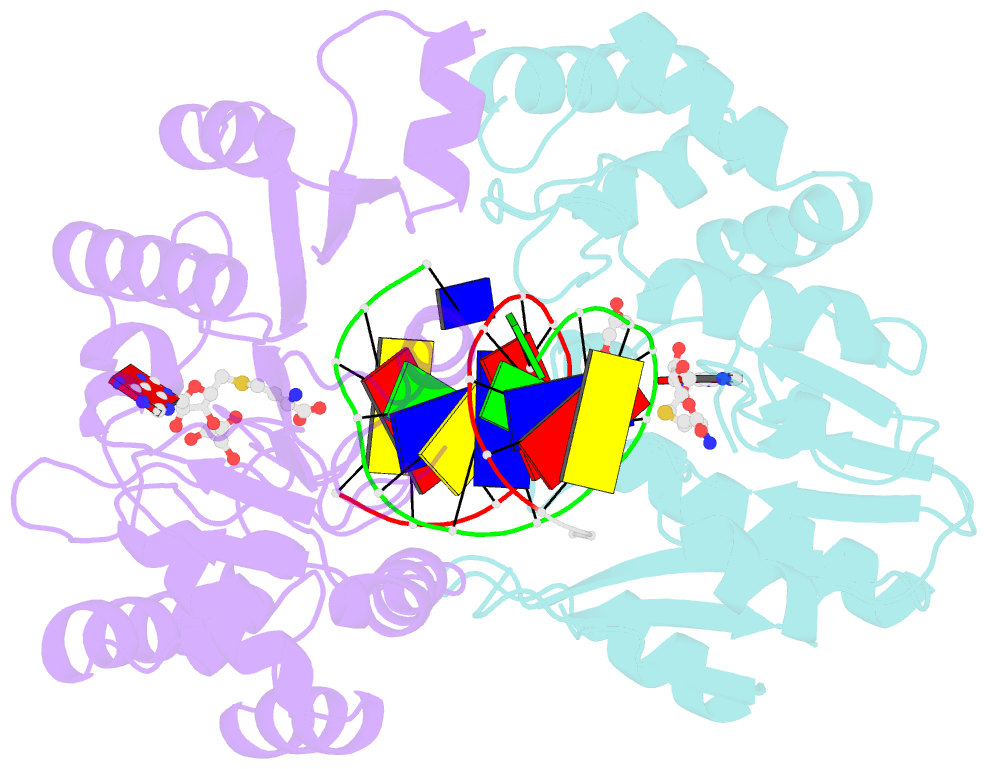
Summary information and primary citation
- PDB-id
- 1yf3; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transferase-DNA
- Method
- X-ray (2.29 Å)
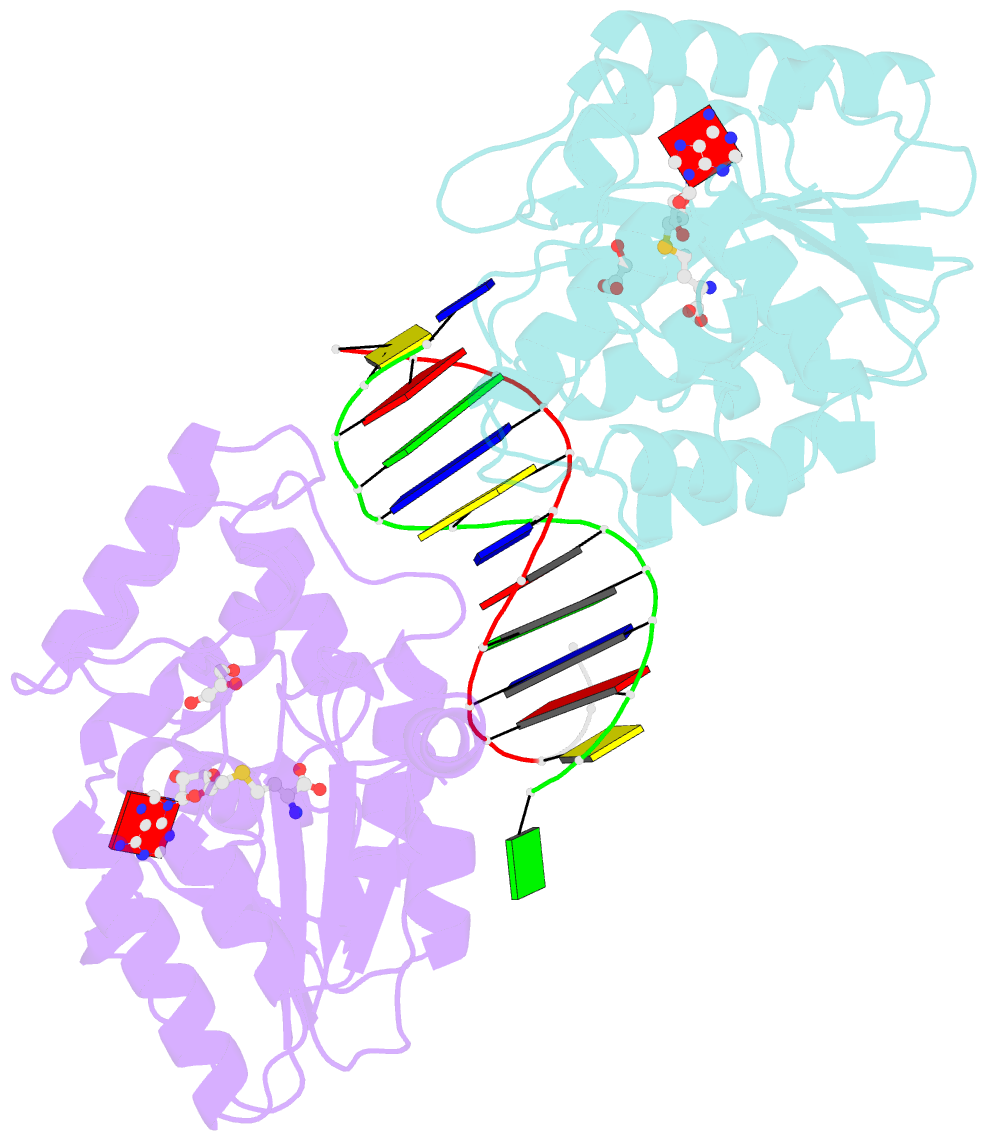
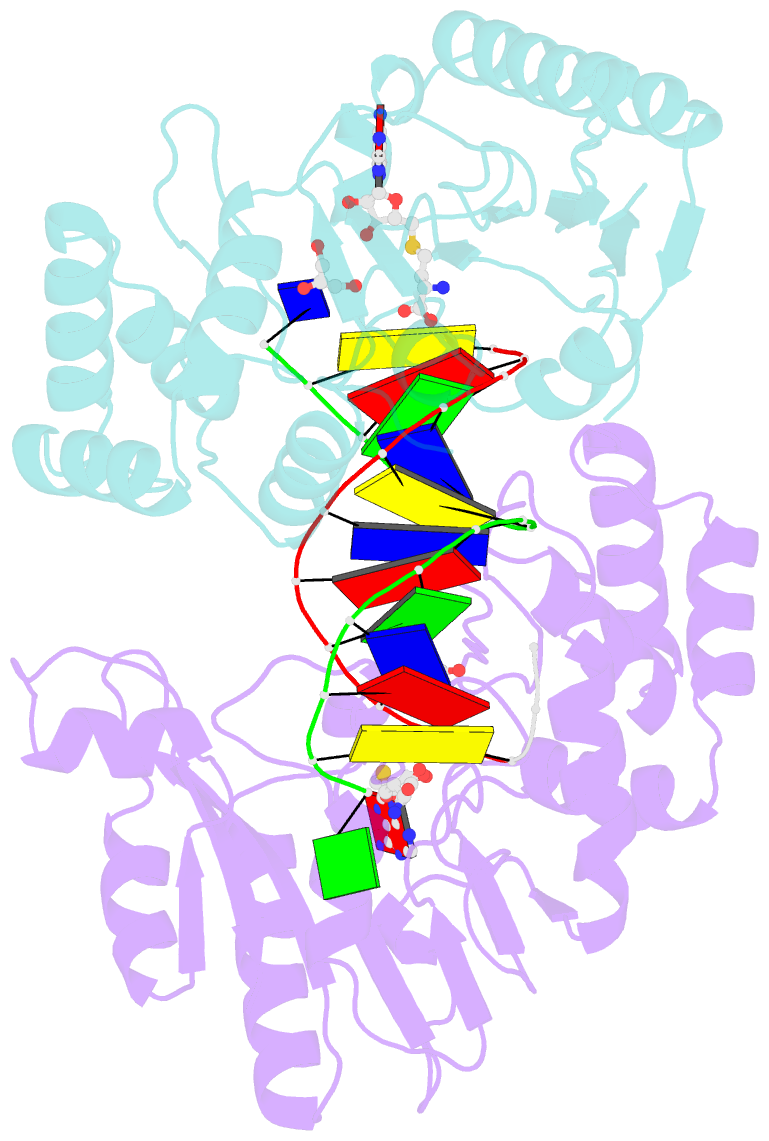
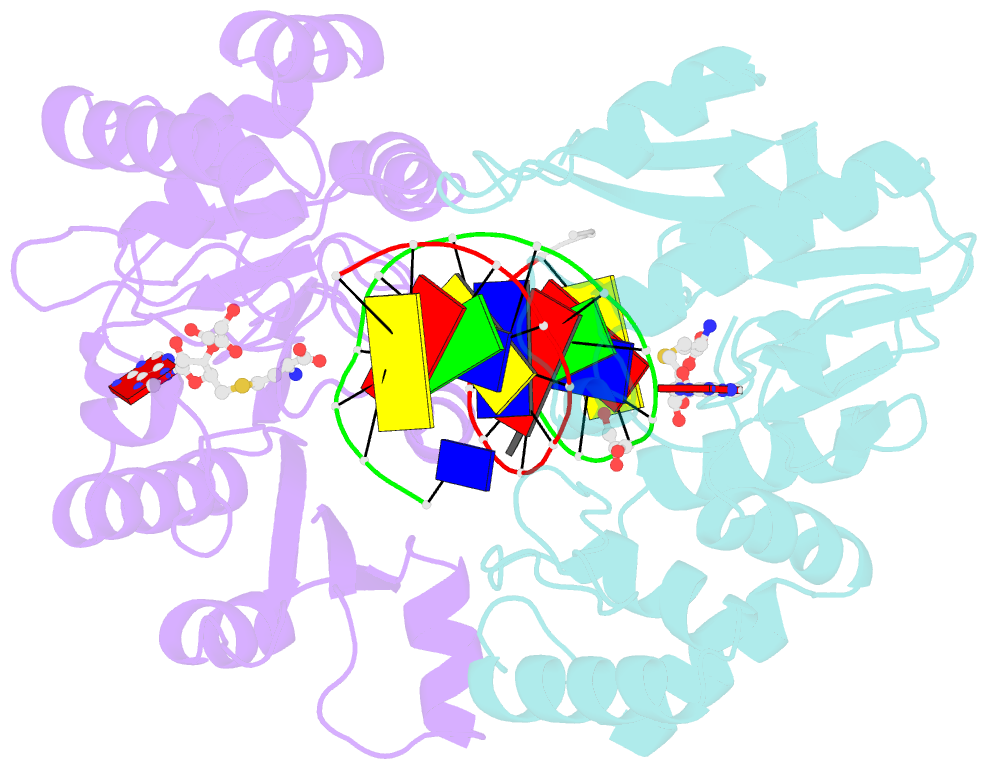
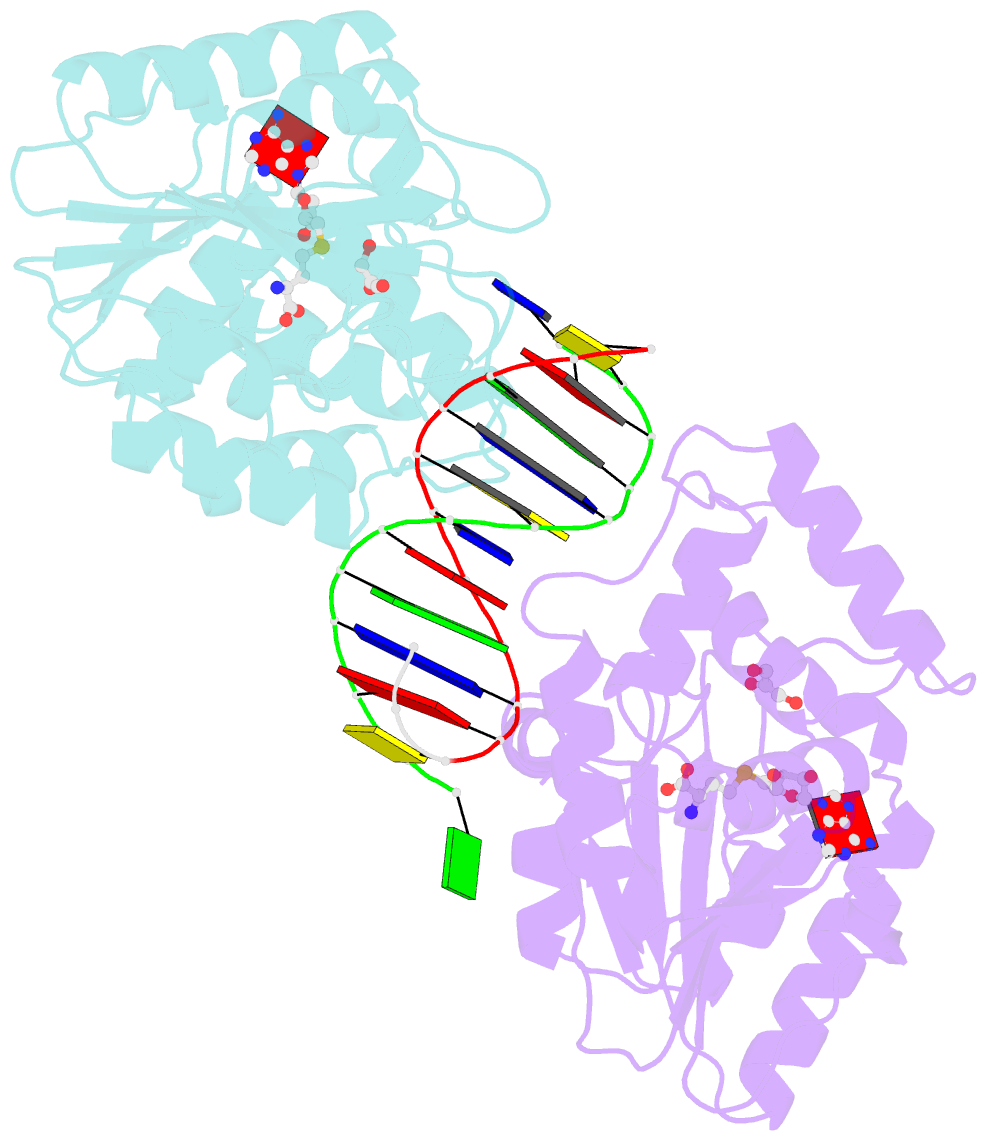
- Summary
- T4dam in complex with adohcy and 13-mer oligonucleotide making non- and semi-specific (~1-4) contact
- Reference
- Horton JR, Liebert K, Hattman S, Jeltsch A, Cheng X (2005): "Transition from Nonspecific to Specific DNA Interactions along the Substrate-Recognition Pathway of Dam Methyltransferase." Cell(Cambridge,Mass.), 121, 349-361. doi: 10.1016/j.cell.2005.02.021.
- Abstract
- DNA methyltransferases methylate target bases within specific nucleotide sequences. Three structures are described for bacteriophage T4 DNA-adenine methyltransferase (T4Dam) in ternary complexes with partially and fully specific DNA and a methyl-donor analog. We also report the effects of substitutions in the related Escherichia coli DNA methyltransferase (EcoDam), altering residues corresponding to those involved in specific interaction with the canonical GATC target sequence in T4Dam. We have identified two types of protein-DNA interactions: discriminatory contacts, which stabilize the transition state and accelerate methylation of the cognate site, and antidiscriminatory contacts, which do not significantly affect methylation of the cognate site but disfavor activity at noncognate sites. These structures illustrate the transition in enzyme-DNA interaction from nonspecific to specific interaction, suggesting that there is a temporal order for formation of specific contacts.