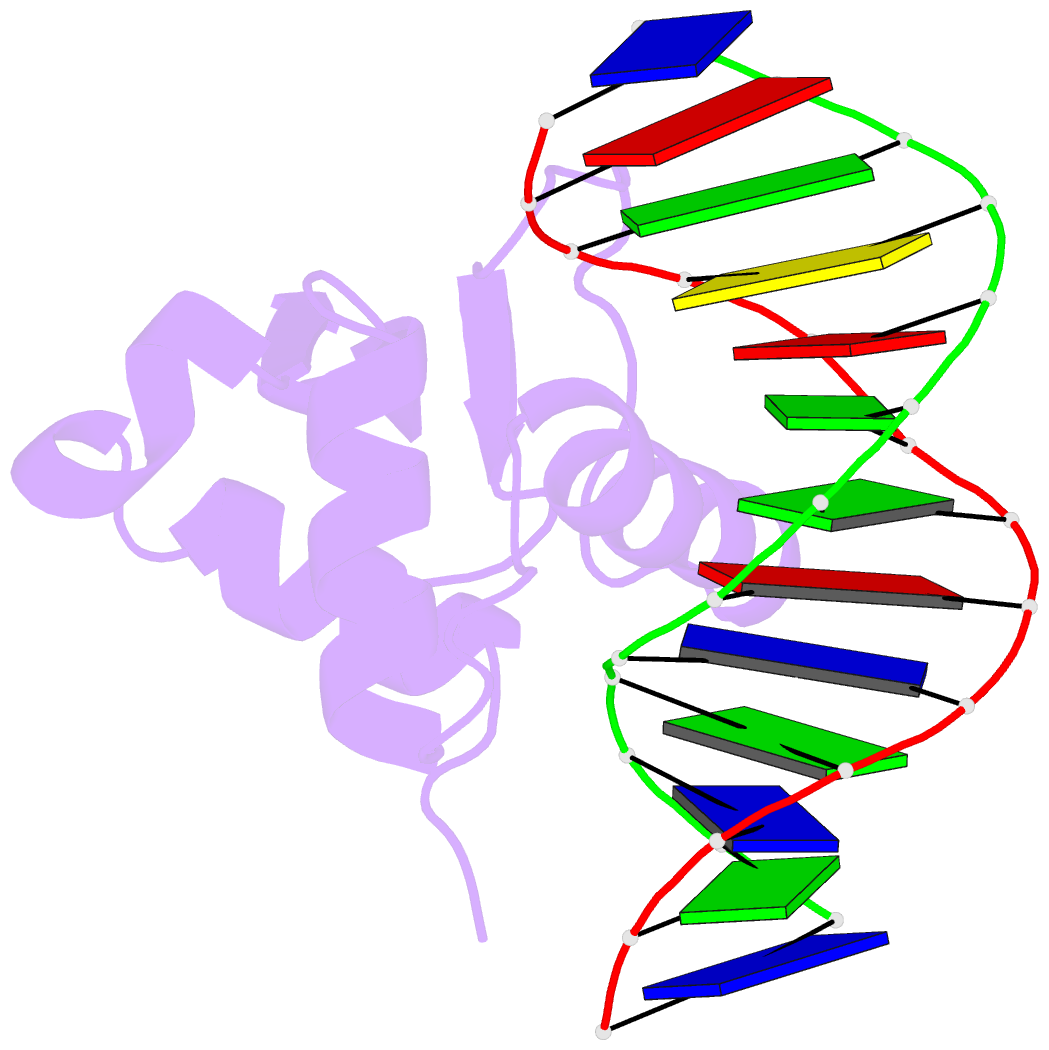
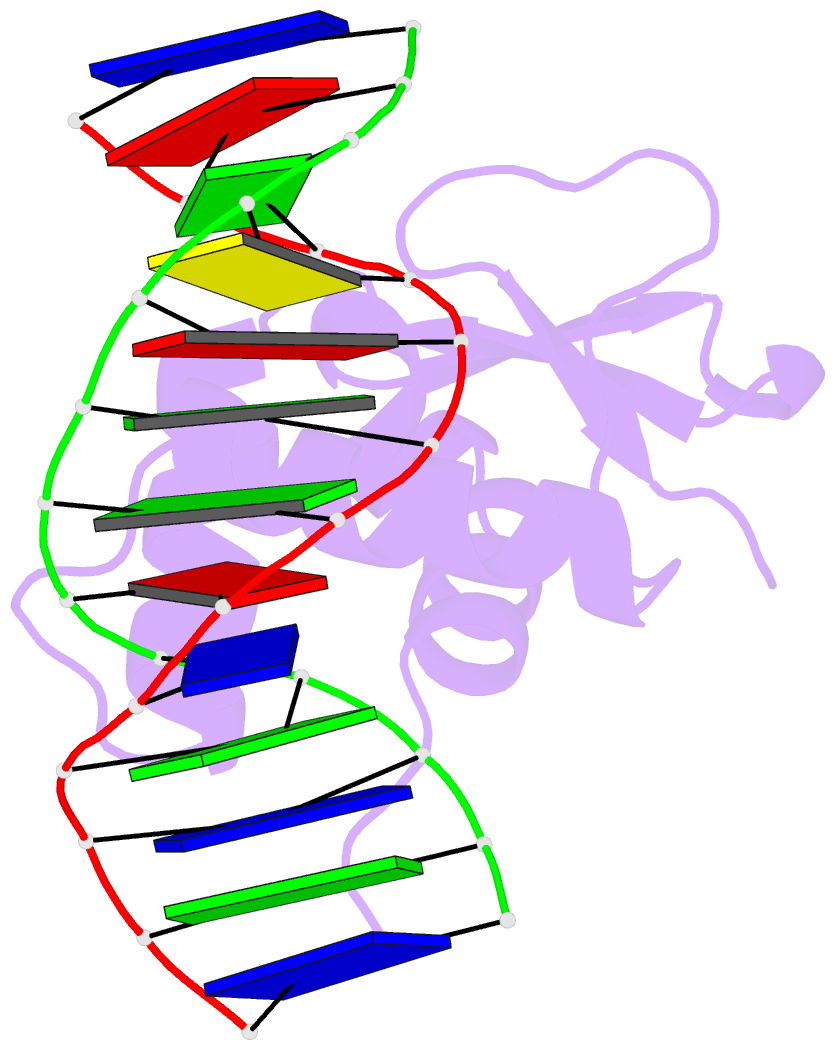
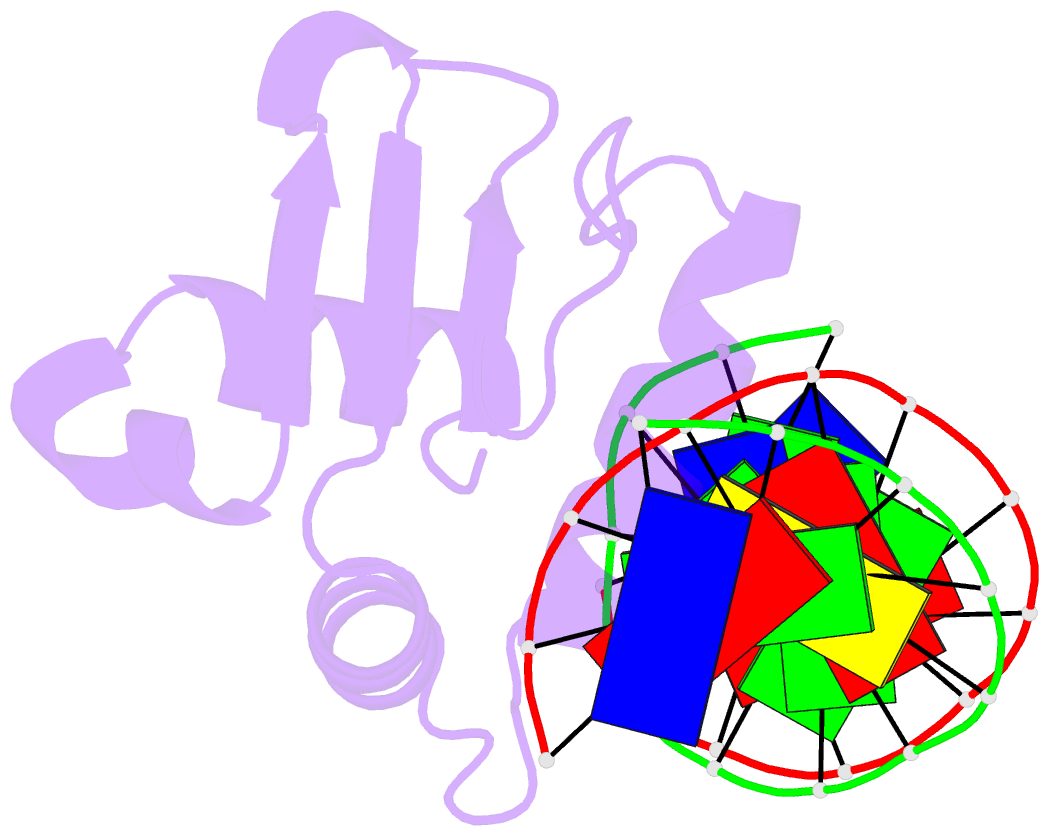
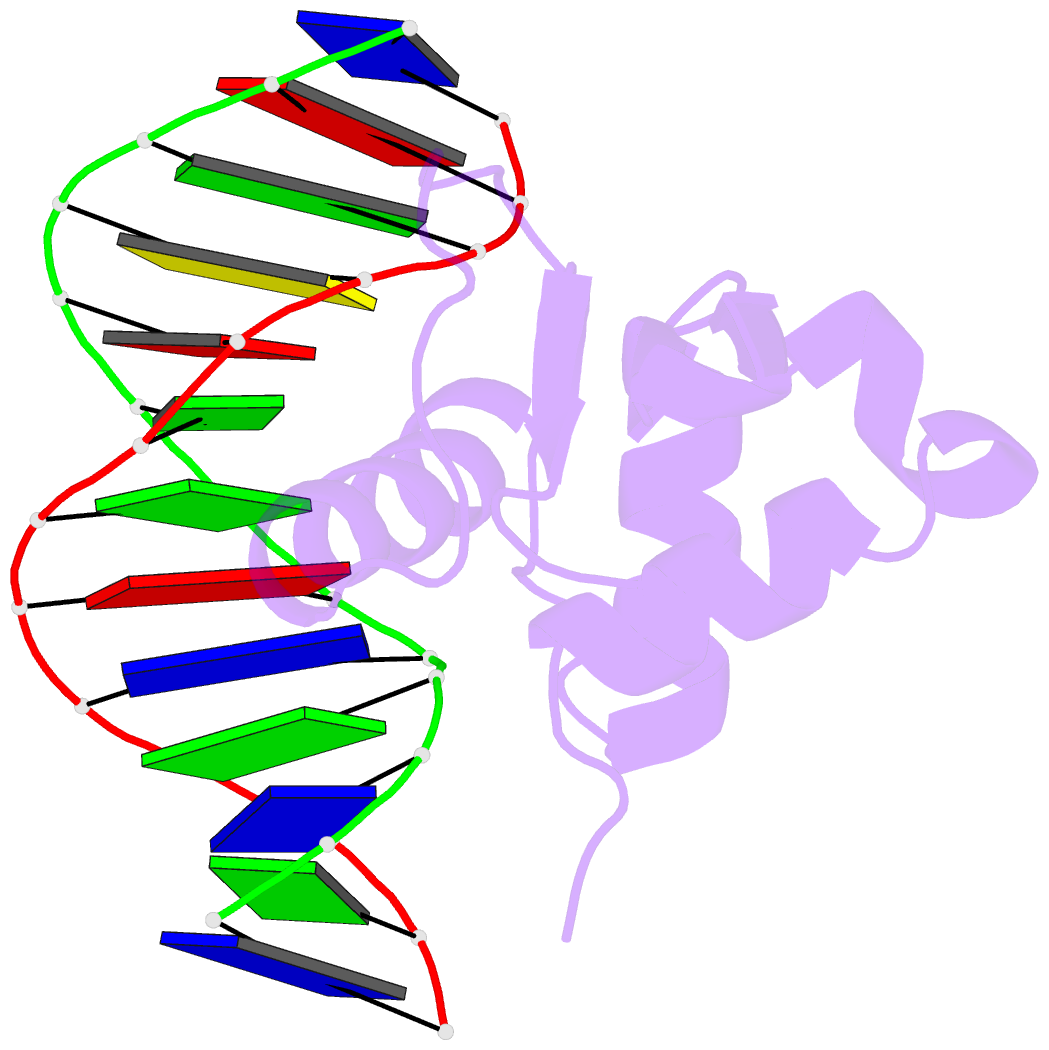
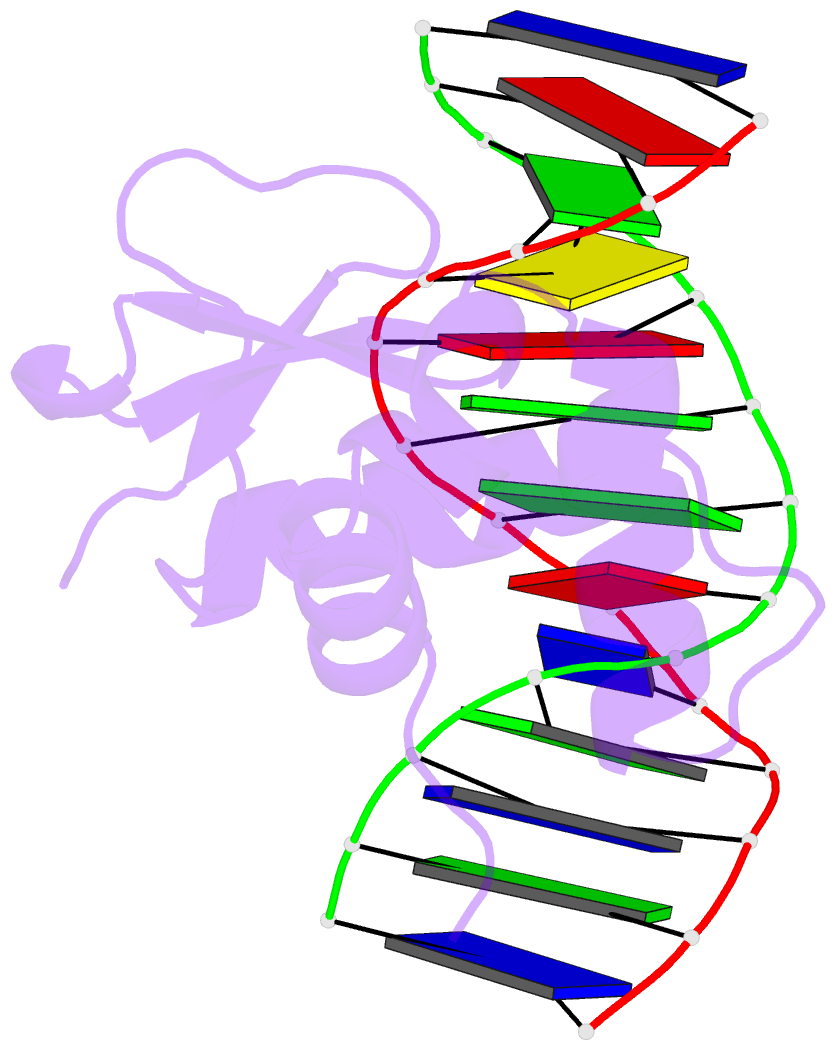
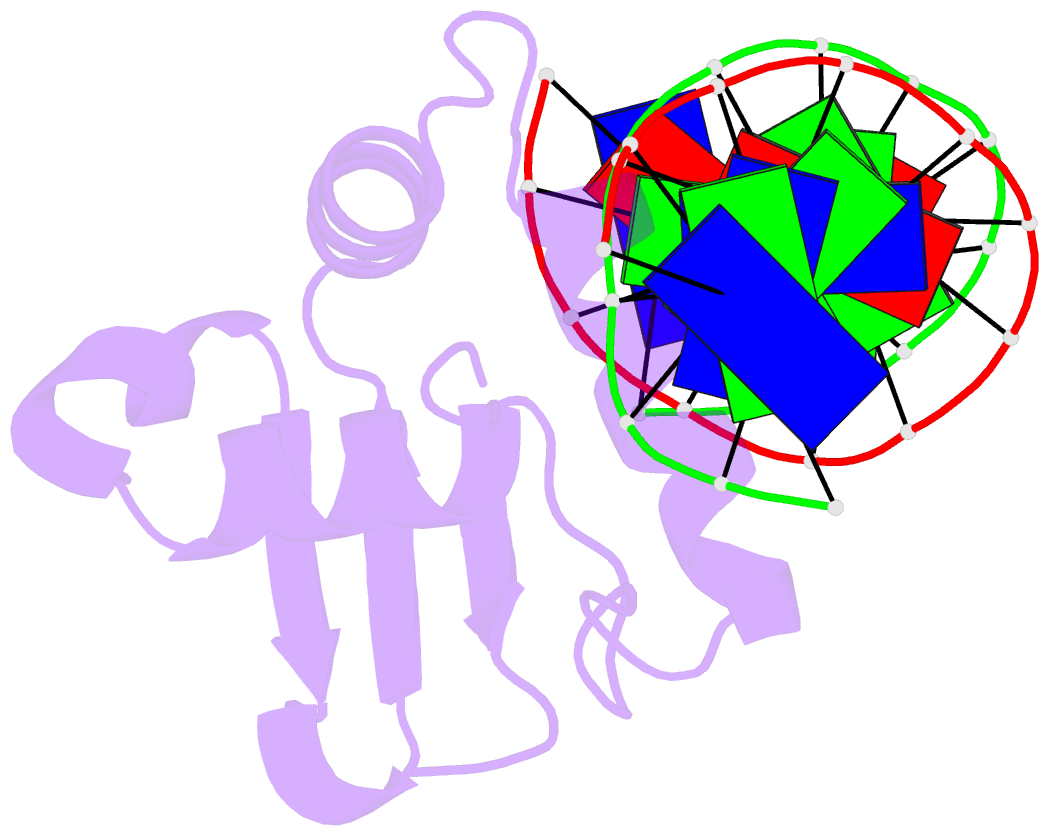
Summary information and primary citation
- PDB-id
- 1yo5; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transcription-DNA
- Method
- X-ray (2.0 Å)
- Summary
- Analysis of the 2.0a crystal structure of the protein-DNA complex of human pdef ets domain bound to the prostate specific antigen regulatory site
- Reference
- Wang Y, Feng L, Said M, Balderman S, Fayazi Z, Liu Y, Ghosh D, Gulick AM (2005): "Analysis of the 2.0 A Crystal Structure of the Protein-DNA Complex of the Human PDEF Ets Domain Bound to the Prostate Specific Antigen Regulatory Site." Biochemistry, 44, 7095-7106. doi: 10.1021/bi047352t.
- Abstract
- PDEF, a prostate epithelial specific transcription factor, is a member of the Ets family of DNA binding proteins. Here we report a 2.0 A crystal structure of the PDEF Ets domain in complex with a natural, high-affinity DNA binding site in the promoter/enhancer region of the human prostate specific antigen gene. Comparison of the PDEF-DNA complex with other Ets complexes revealed key features that are shared among Ets members, as well as important differences in substrate specification at both the "GGA" core and the flanking regions of the DNA site. The combination of the serine residue at position 308 and the glutamine at position 311 explains the previous observation that the PDEF binds preferentially to a thymine at the +4 position of its binding site. Despite the common essential features that are shared among Ets members, PDEF demonstrates distinct patterns of interactions at different positions of DNA in achieving sequence specific recognition. Collectively, the common and unique interactions with both the DNA bases and the backbone phosphates lead to substrate specificity and individual preference for certain DNA sites.