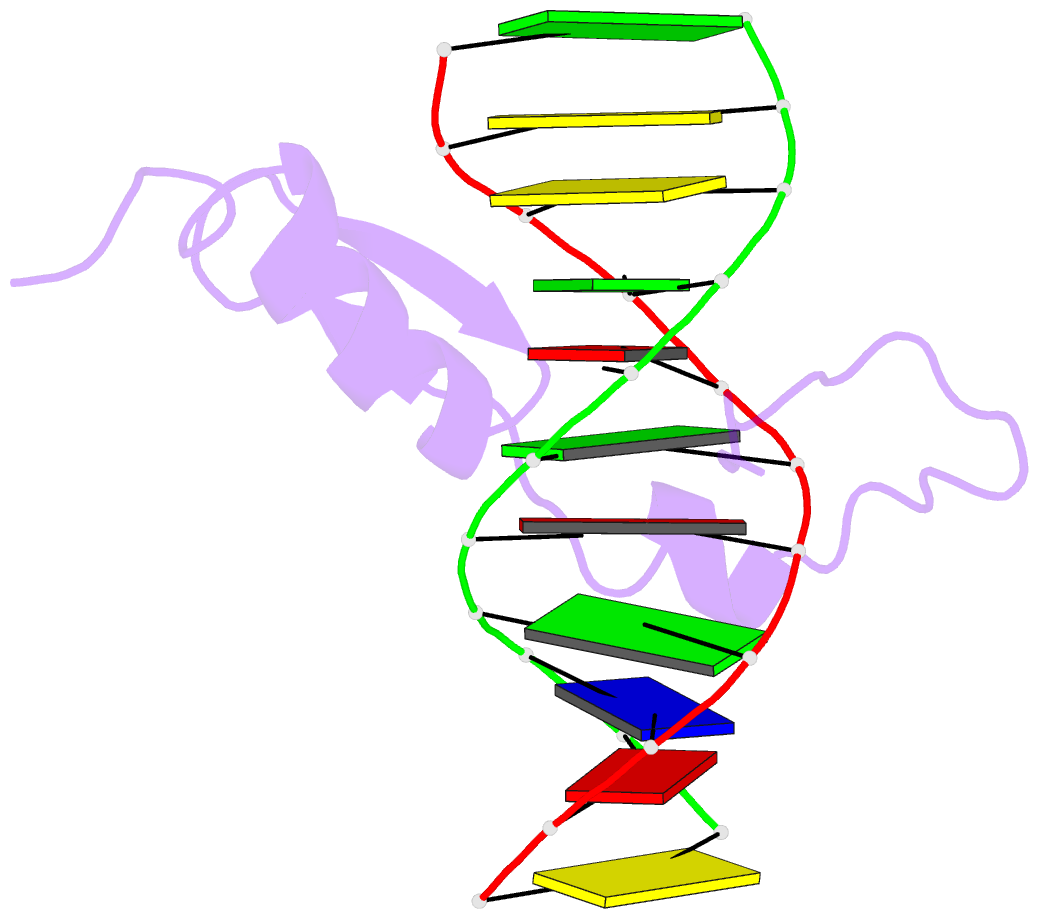
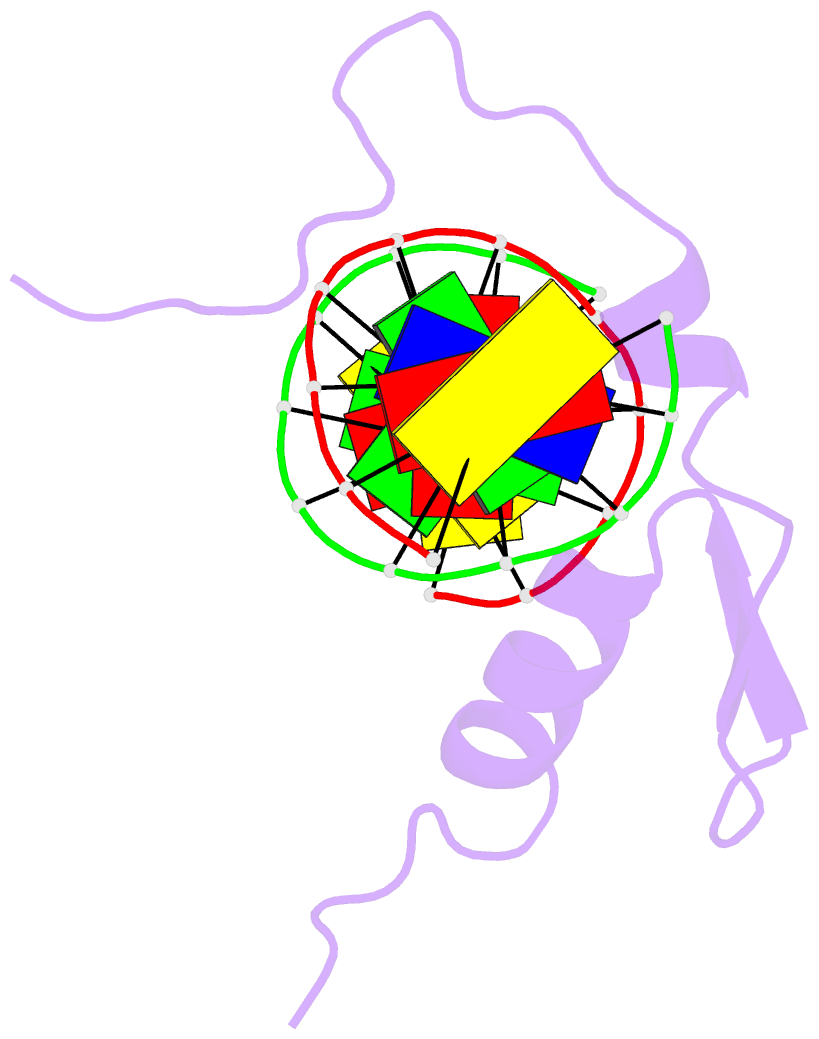
Summary information and primary citation
- PDB-id
- 1yui; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- DNA binding protein-DNA
- Method
- NMR
- Summary
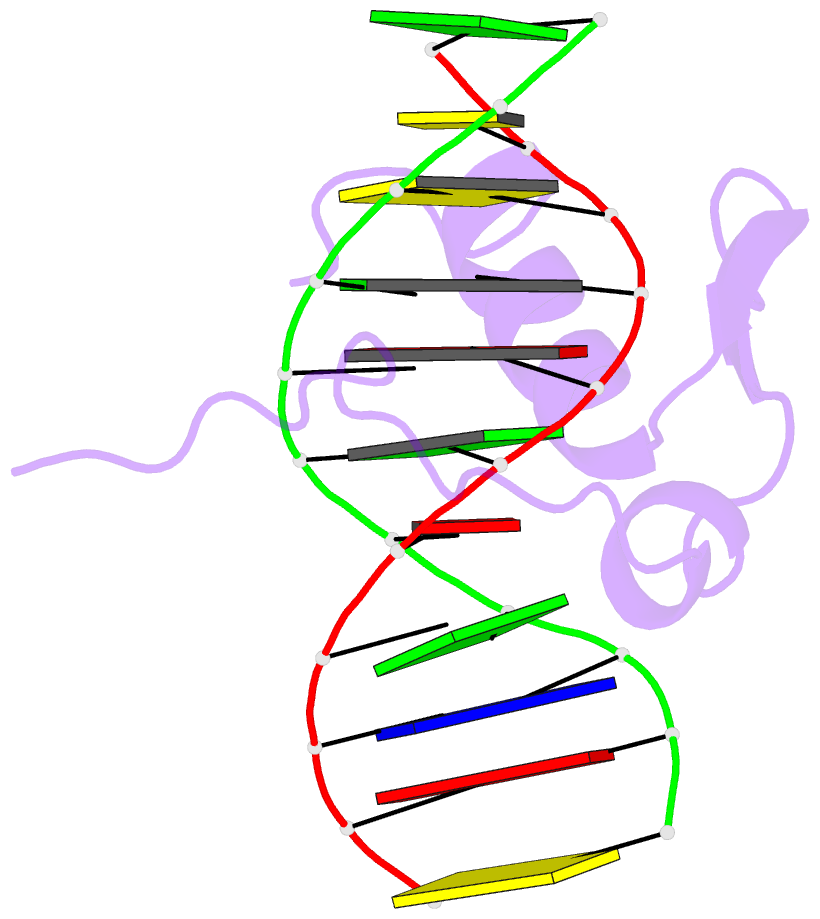
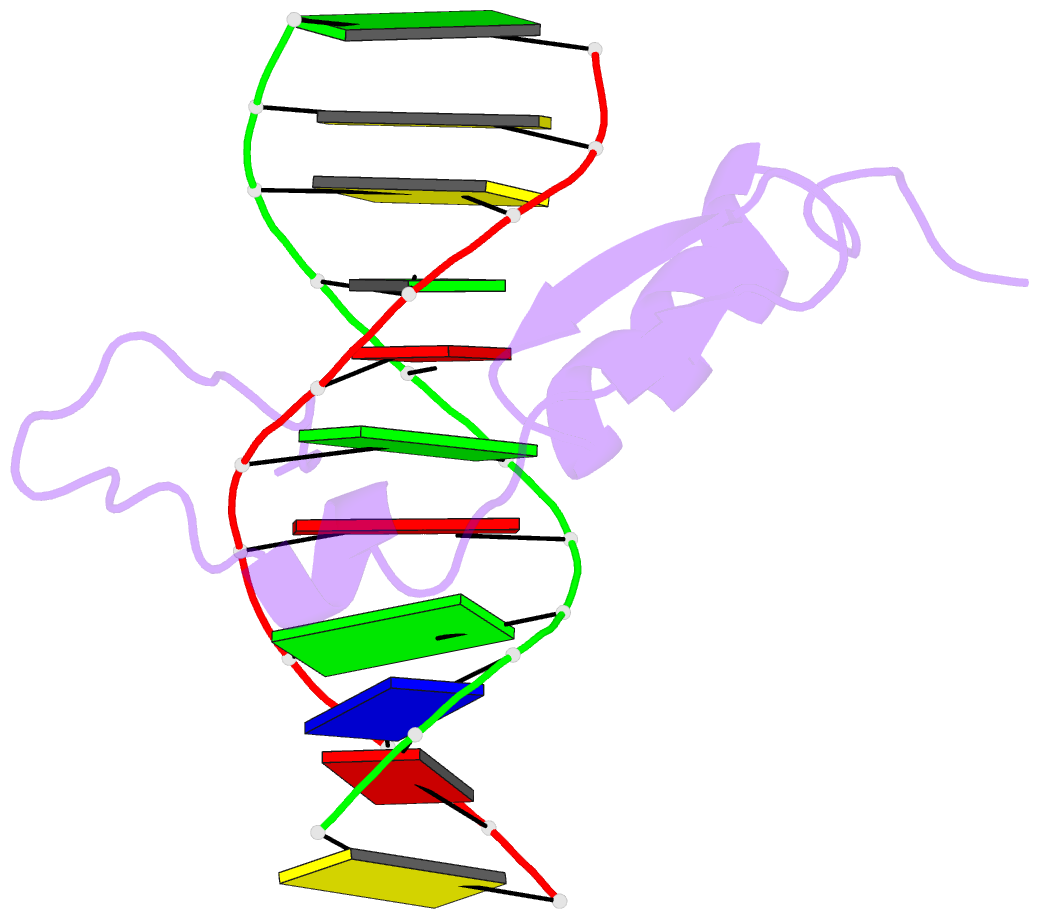
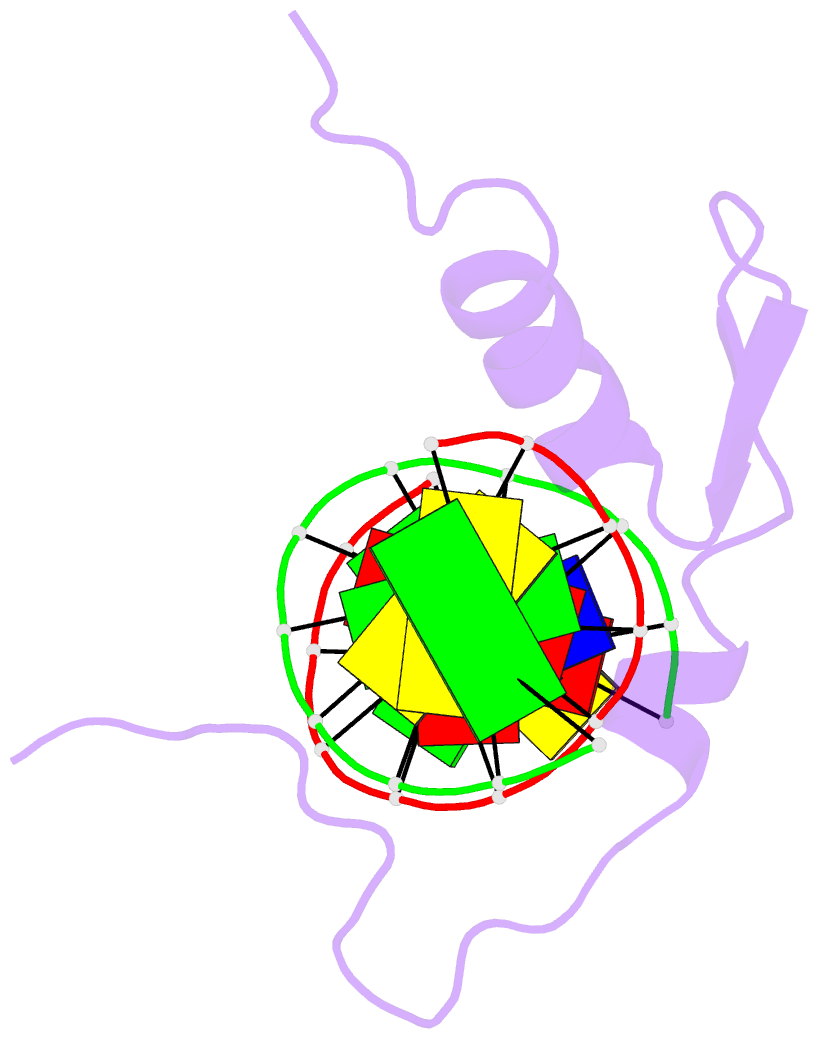
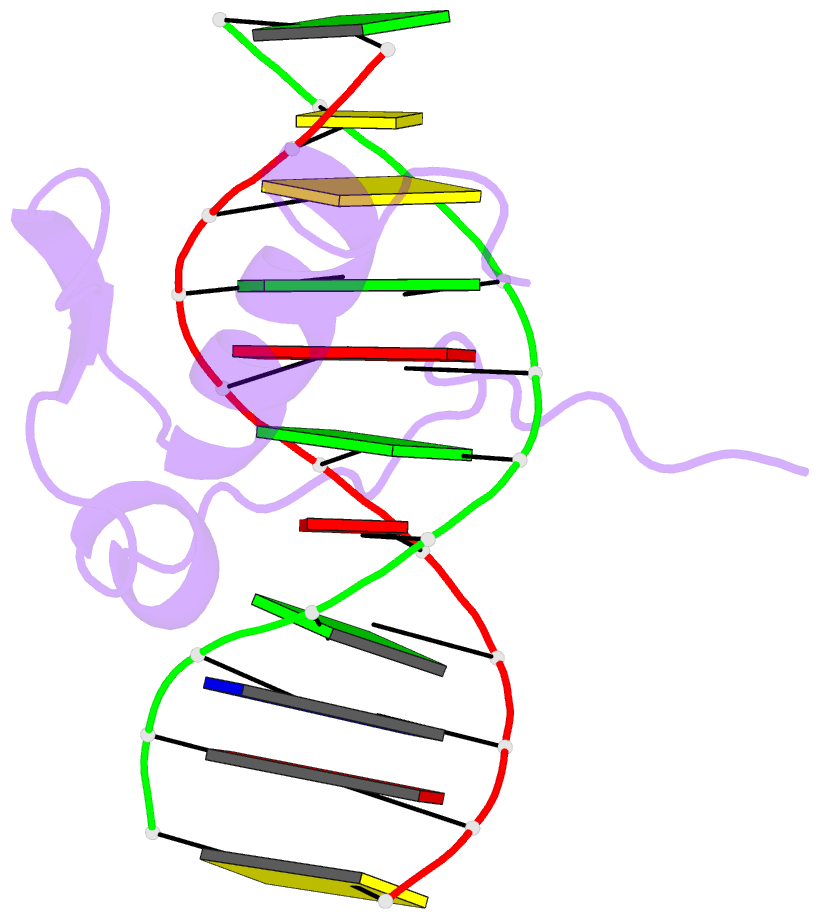
- Solution NMR structure of the gaga factor-DNA complex, regularized mean structure
- Reference
- Omichinski JG, Pedone PV, Felsenfeld G, Gronenborn AM, Clore GM (1997): "The solution structure of a specific GAGA factor-DNA complex reveals a modular binding mode." Nat.Struct.Biol., 4, 122-132. doi: 10.1038/nsb0297-122.
- Abstract
- The structure of a complex between the DNA binding domain of the GAGA factor (GAGA-DBD) and an oligonucleotide containing its GAGAG consensus binding site has been determined by nuclear magnetic resonance spectroscopy. The GAGA-DBD comprises a single classical Cys2-His2 zinc finger core, and an N-terminal extension containing two highly basic regions, BR1 and BR2. The zinc finger core binds in the major groove and recognizes the first three GAG bases of the consensus in a manner similar to that seen in other classical zinc finger-DNA complexes. Unlike the latter, which require tandem zinc finger repeats with a minimum of two units for high affinity binding, the GAGA-DBD makes use of only a single finger complemented by BR1 and BR2. BR2 forms a helix that interacts in the major groove recognizing the last G of the consensus, while BR1 wraps around the DNA in the minor groove and recognizes the A in the fourth position of the consensus. The implications of the structure of the GAGA-DBD-DNA complex for chromatin remodelling are discussed.