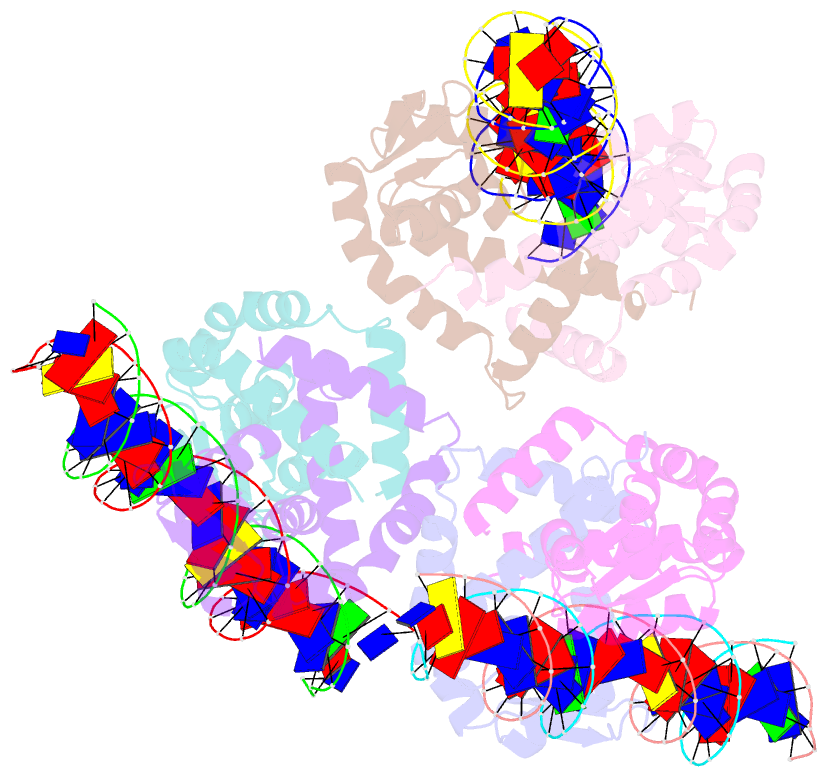
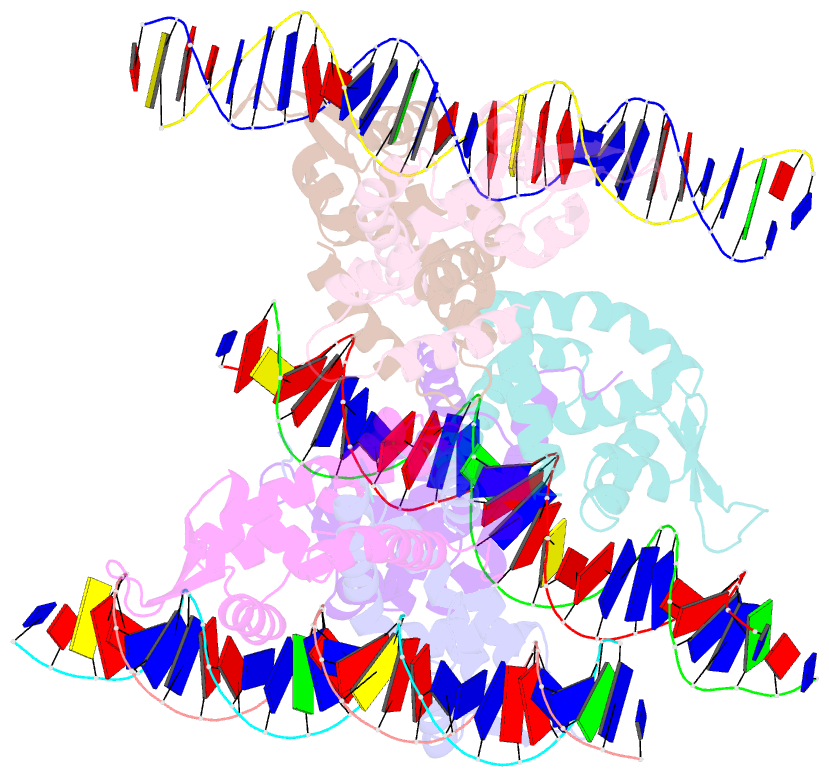
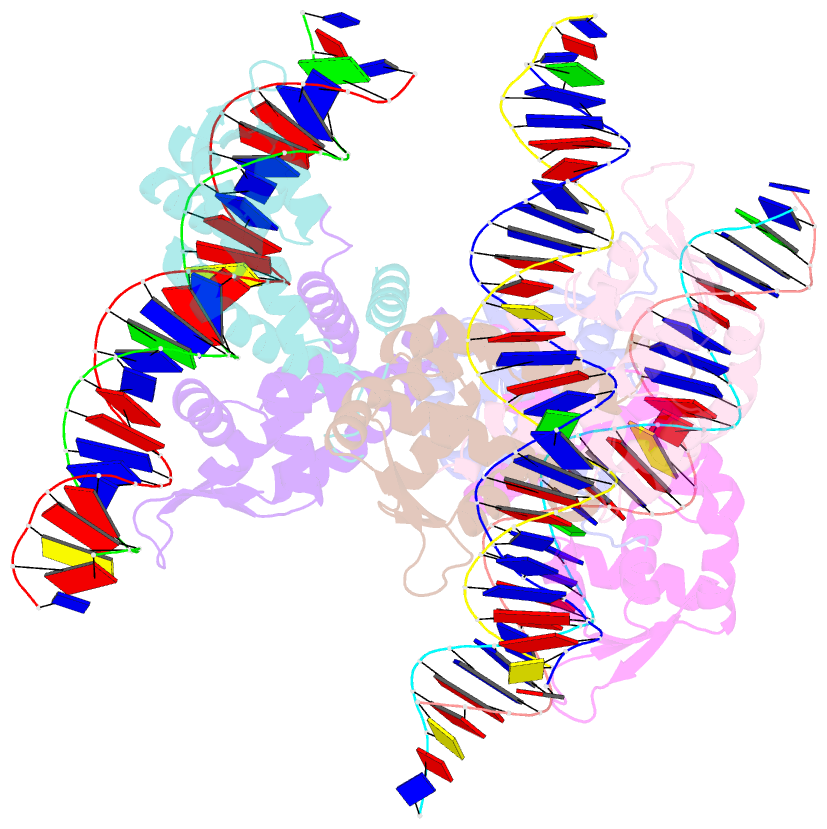
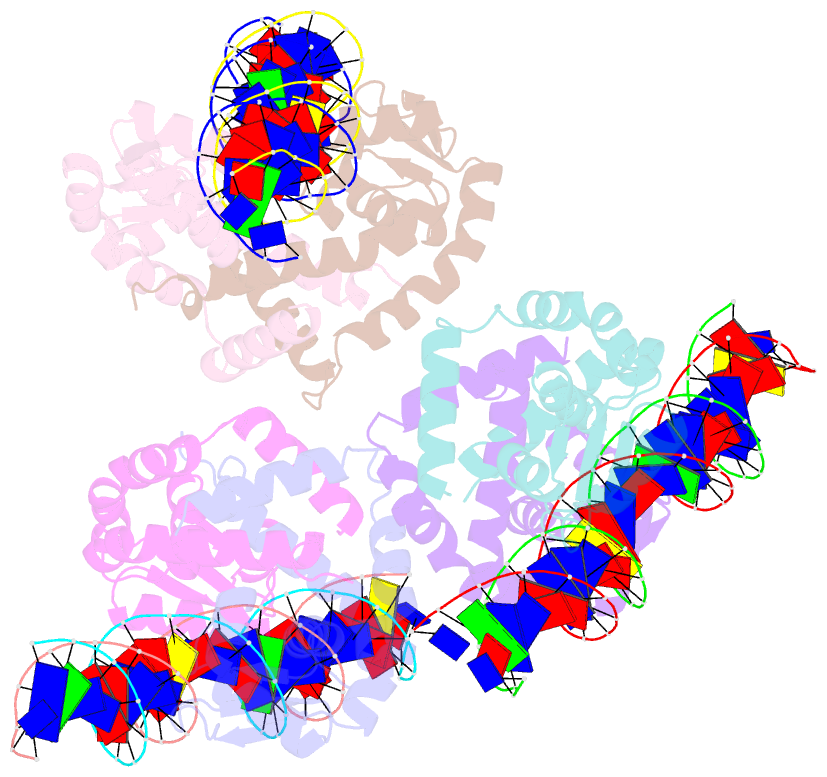
Summary information and primary citation
- PDB-id
- 1z9c; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transcription-DNA
- Method
- X-ray (2.64 Å)
- Summary
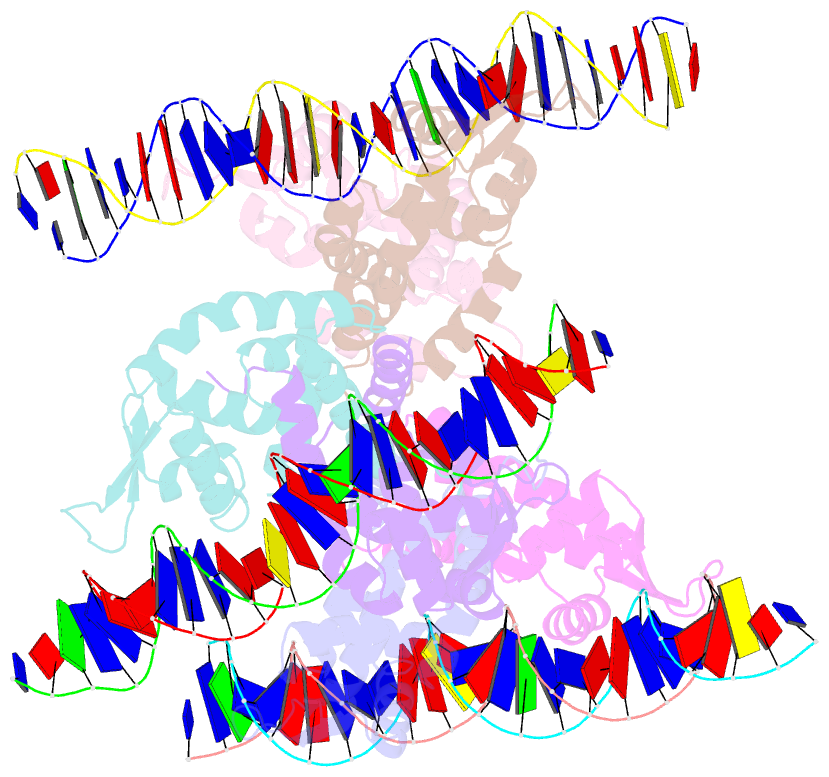
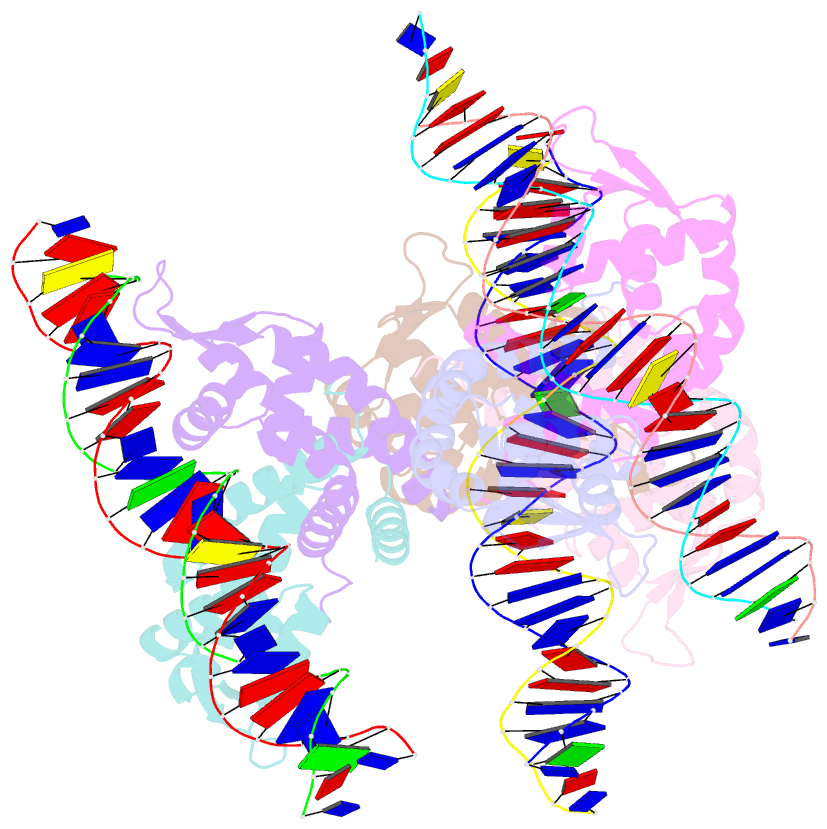
- Crystal structure of ohrr bound to the ohra promoter: structure of marr family protein with operator DNA
- Reference
- Hong M, Fuangthong M, Helmann JD, Brennan RG (2005): "Structure of an OhrR-ohrA Operator Complex Reveals the DNA Binding Mechanism of the MarR Family." Mol.Cell, 20, 131-141. doi: 10.1016/j.molcel.2005.09.013.
- Abstract
- The mechanisms by which Bacillus subtilis OhrR, a member of the MarR family of transcription regulators, binds the ohrA operator and is induced by oxidation of its lone cysteine residue by organic hydroperoxides to sulphenic acid are unknown. Here, we describe the crystal structures of reduced OhrR and an OhrR-ohrA operator complex. To bind DNA, OhrR employs a chimeric winged helix-turn-helix DNA binding motif, which is composed of extended eukaryotic-like wings, prokaryotic helix-turn-helix motifs, and helix-helix elements. The reactivity of the peroxide-sensing cysteine is not modulated by proximal basic residues but largely by the positive dipole of helix alpha1. Induction originates from the alleviation of intersubunit steric clash between the sulphenic acid moieties of the oxidized sensor cysteines and nearby tyrosines and methionines. The structure of the OhrR-ohrA operator complex reveals the DNA binding mechanism of the entire MarR family and suggests a common inducer binding pocket.