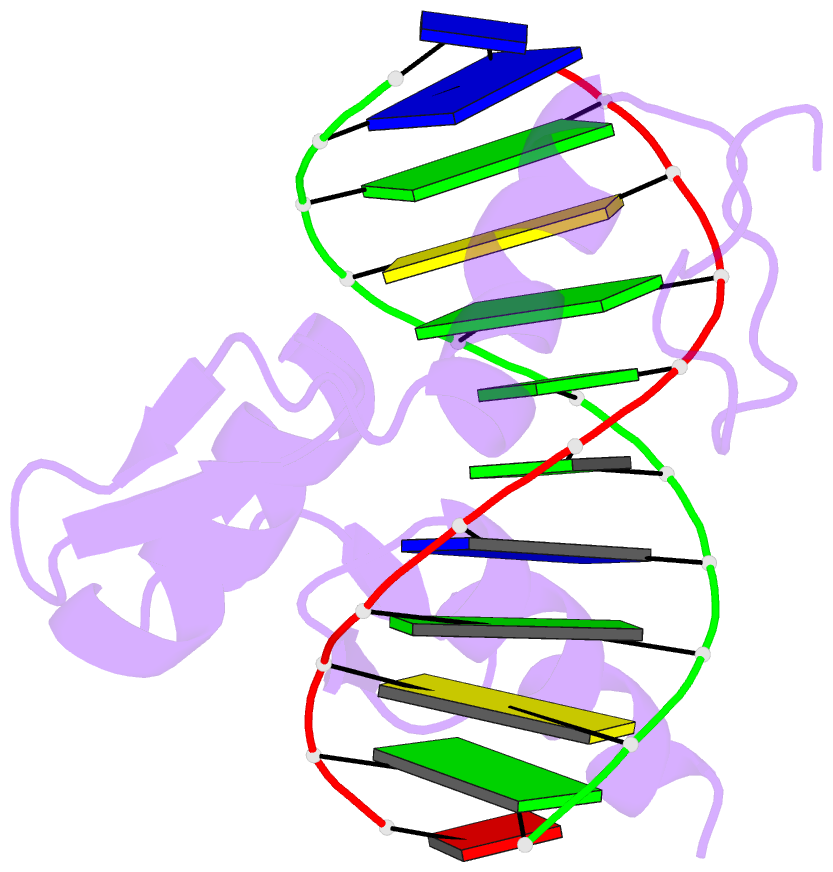
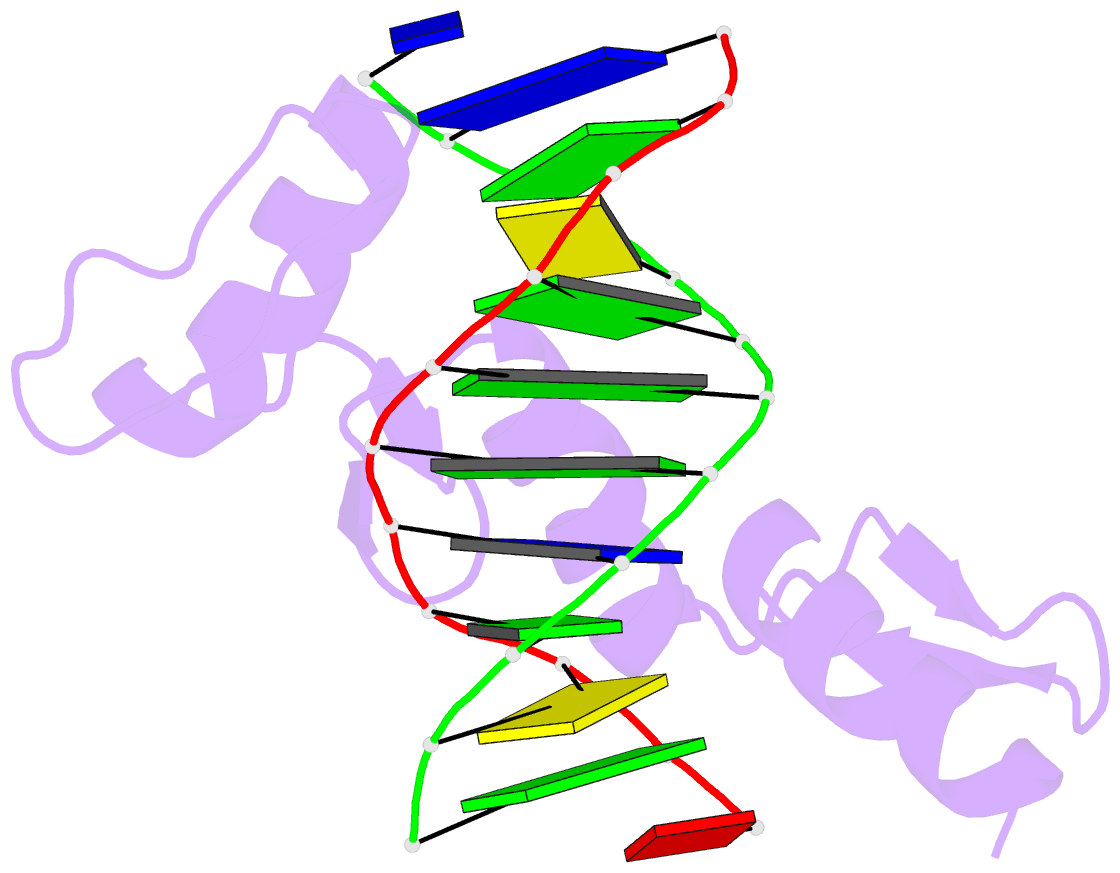
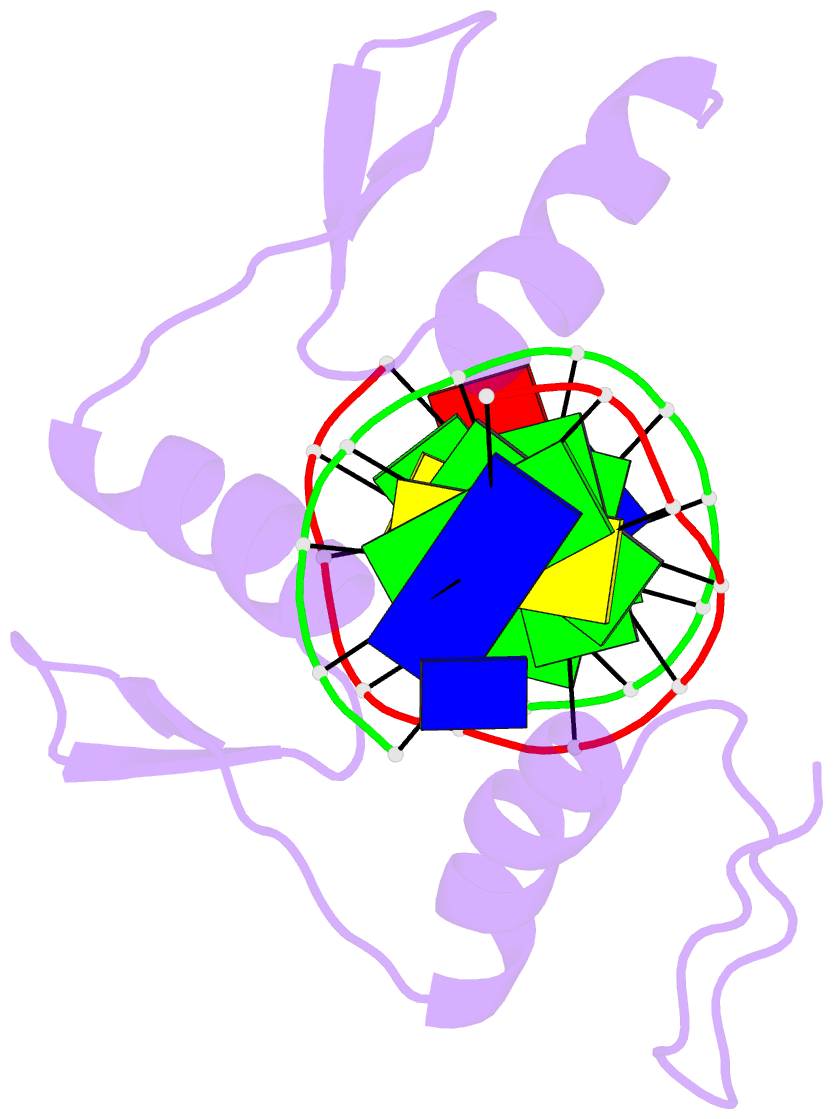
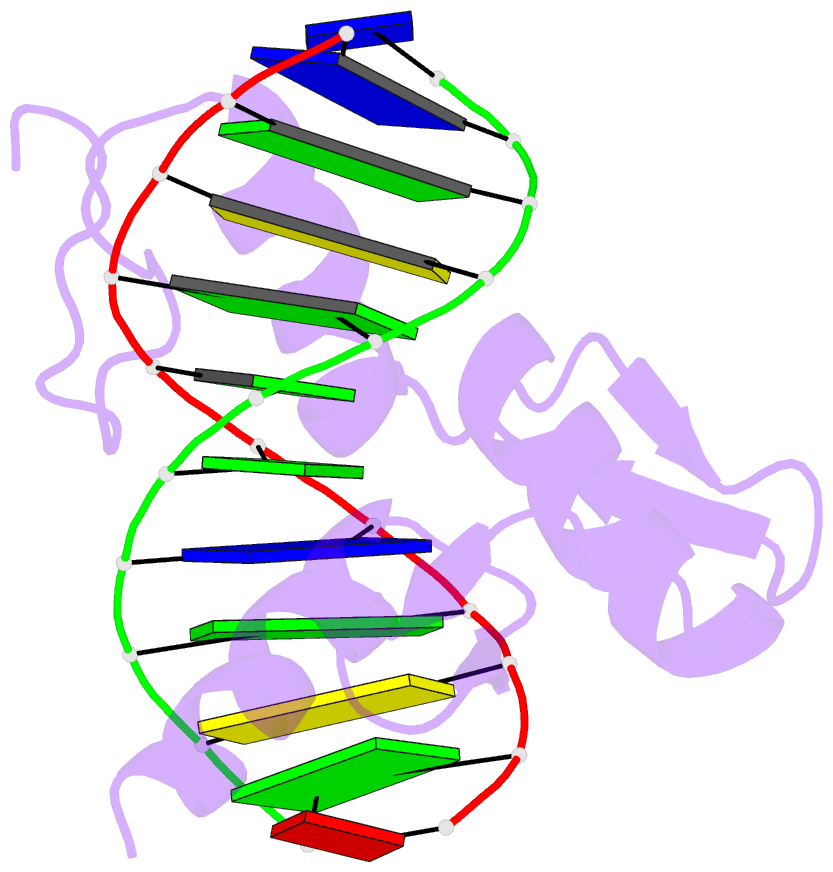
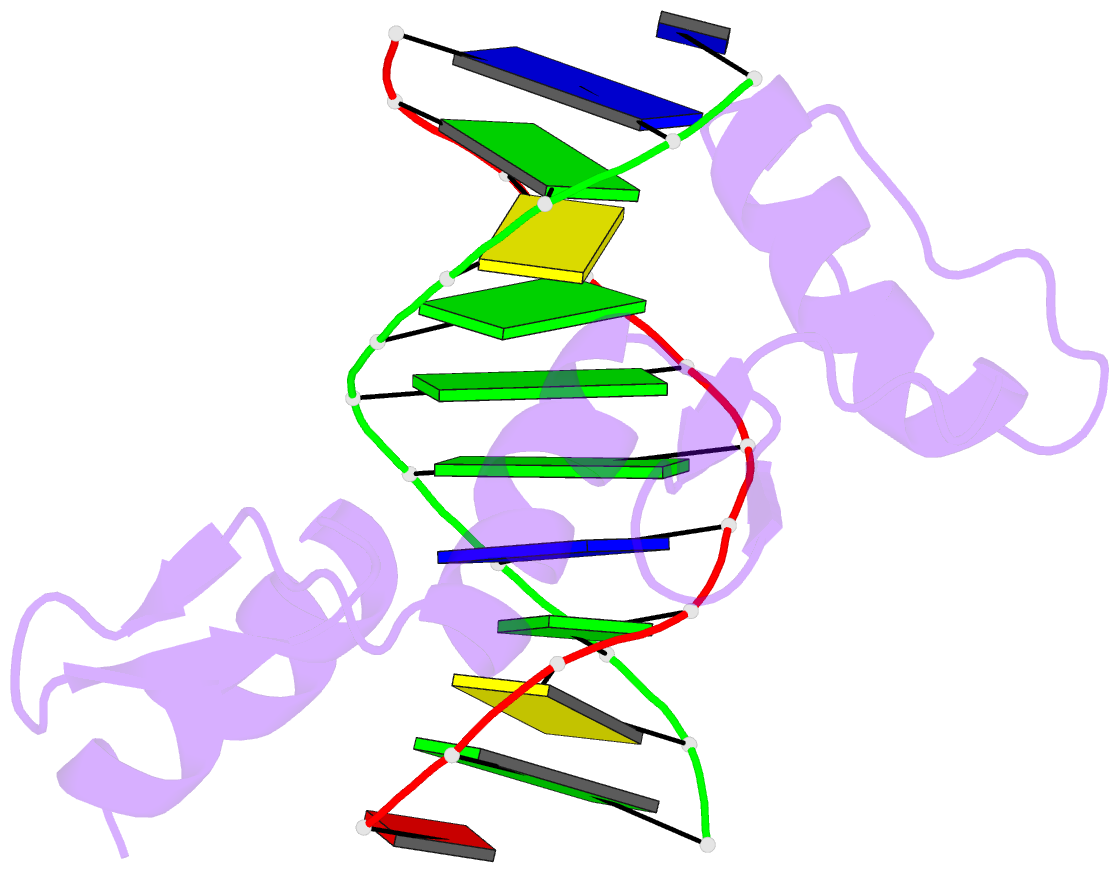
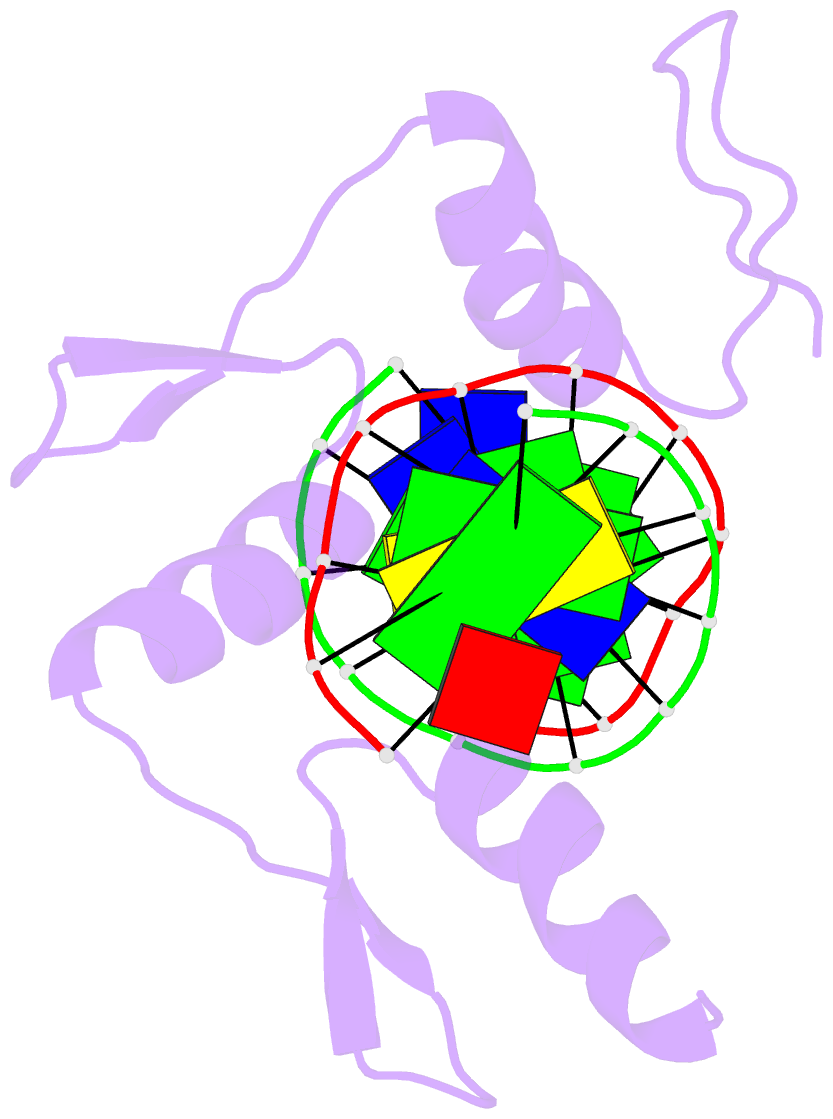
Summary information and primary citation
- PDB-id
- 1zaa; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transcription-DNA
- Method
- X-ray (2.1 Å)
- Summary
- Zinc finger-DNA recognition: crystal structure of a zif268-DNA complex at 2.1 angstroms
- Reference
- Pavletich NP, Pabo CO (1991): "Zinc finger-DNA recognition: crystal structure of a Zif268-DNA complex at 2.1 A." Science, 252, 809-817.
- Abstract
- The zinc finger DNA-binding motif occurs in many proteins that regulate eukaryotic gene expression. The crystal structure of a complex containing the three zinc fingers from Zif268 (a mouse immediate early protein) and a consensus DNA-binding site has been determined at 2.1 angstroms resolution and refined to a crystallographic R factor of 18.2 percent. In this complex, the zinc fingers bind in the major groove of B-DNA and wrap part way around the double helix. Each finger has a similar relation to the DNA and makes its primary contacts in a three-base pair subsite. Residues from the amino-terminal portion of an alpha helix contact the bases, and most of the contracts are made with the guanine-rich strand of the DNA. This structure provides a framework for understanding how zinc fingers recognize DNA and suggests that this motif may provide a useful basis for the design of novel DNA-binding proteins.