Summary information and primary citation
- PDB-id
- 1zbn; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- RNA binding protein-RNA
- Method
- NMR
- Summary
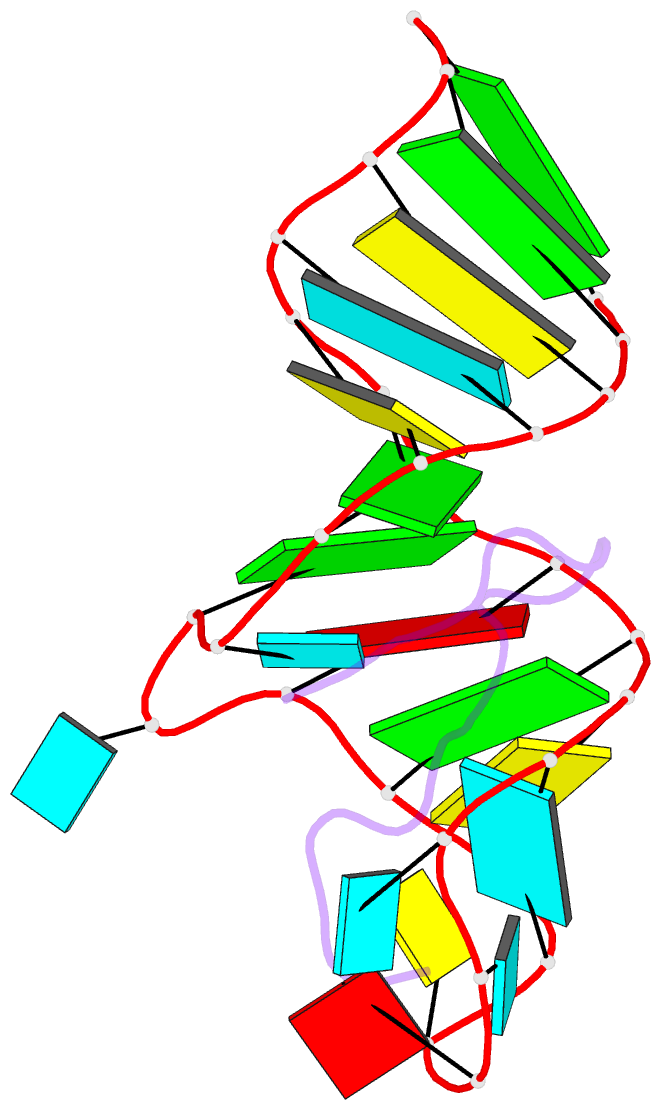
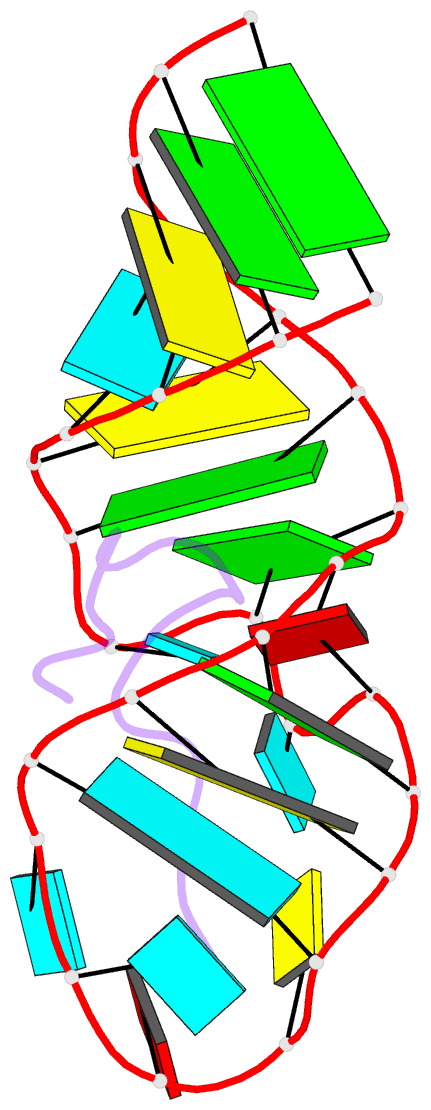
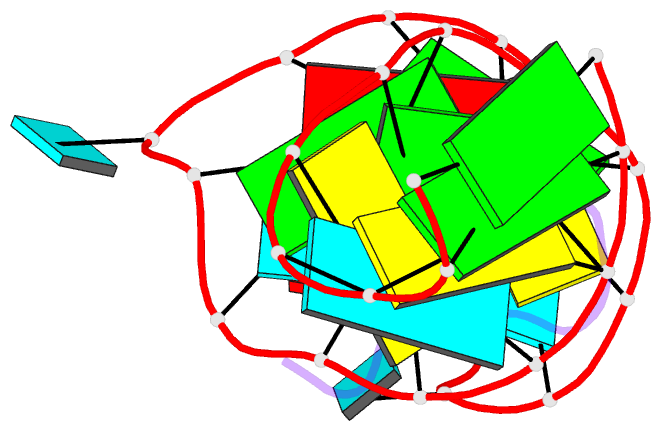
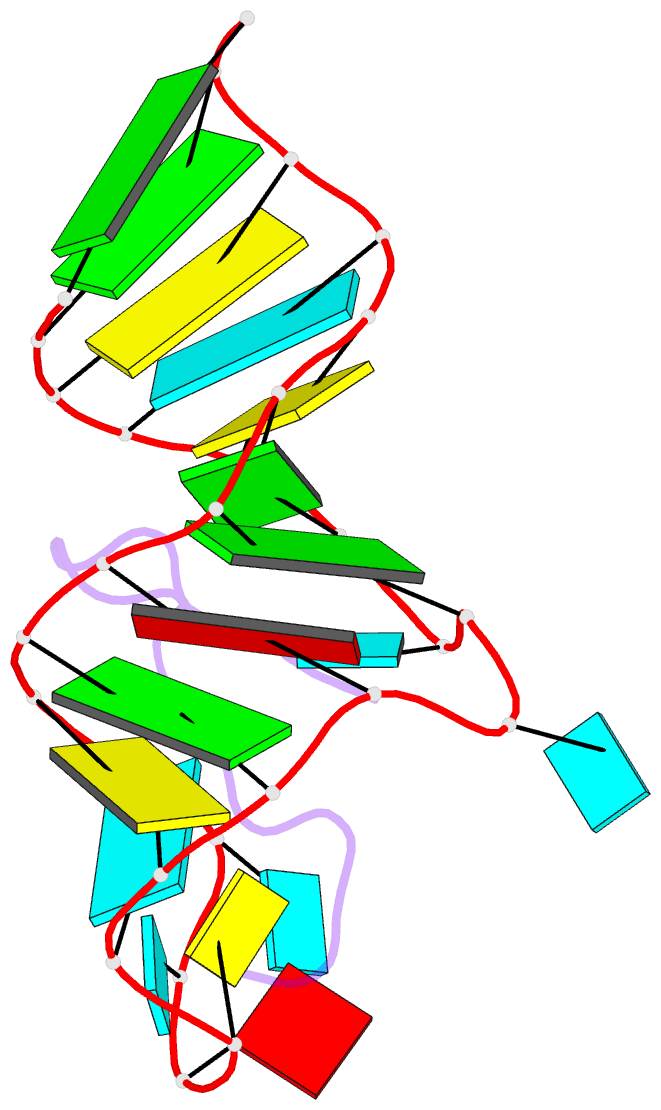
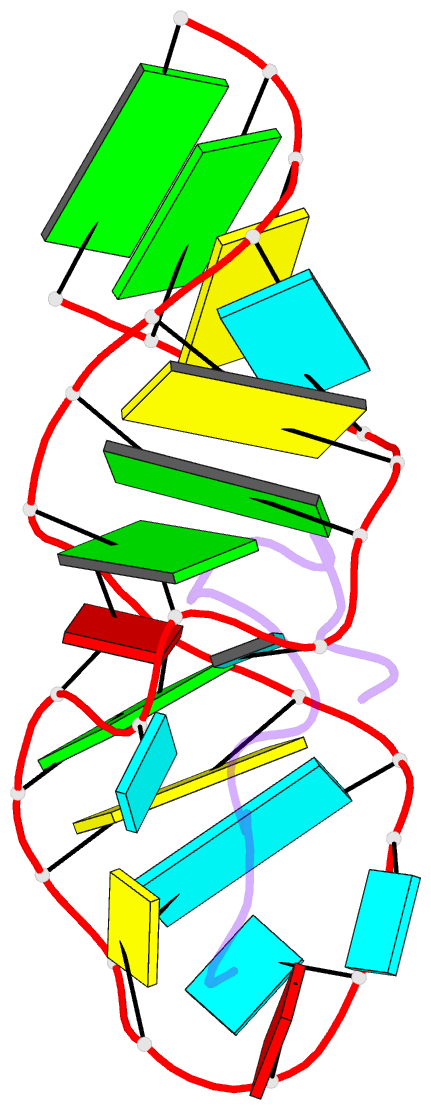
- Solution structure of biv tar hairpin complexed to jdv tat arginine-rich motif
- Reference
- Calabro V, Daugherty MD, Frankel AD (2005): "A single intermolecular contact mediates intramolecular stabilization of both RNA and protein." Proc.Natl.Acad.Sci.Usa, 102, 6849-6854. doi: 10.1073/pnas.0409282102.
- Abstract
- An arginine-rich peptide from the Jembrana disease virus (JDV) Tat protein is a structural "chameleon" that binds bovine immunodeficiency virus (BIV) or HIV TAR RNAs in two different binding modes, with an affinity for BIV TAR even higher than the cognate BIV peptide. We determined the NMR structure of the JDV Tat-BIV TAR high-affinity complex and found that the C-terminal tyrosine in JDV Tat forms a network of inter- and intramolecular hydrogen bonding and stacking interactions that simultaneously stabilize the beta-hairpin conformation of the peptide and a base triple in the RNA. A neighboring histidine also appears to help stabilize the peptide conformation. Induced fit binding is recurrent in protein-protein and protein-nucleic acid interactions, and the JDV Tat complex demonstrates how high affinity can be achieved not only by optimization of the binding interface but also by inducing new intramolecular contacts that stabilize each binding partner. Comparison to the cognate BIV Tat peptide-TAR complex shows how such a costabilization mechanism can evolve with only small changes to the peptide sequence. In addition, the bound structure of BIV TAR in the chameleon peptide complex is strikingly similar to the bound conformation of HIV TAR, suggesting new strategies for the development of HIV TAR binding molecules.