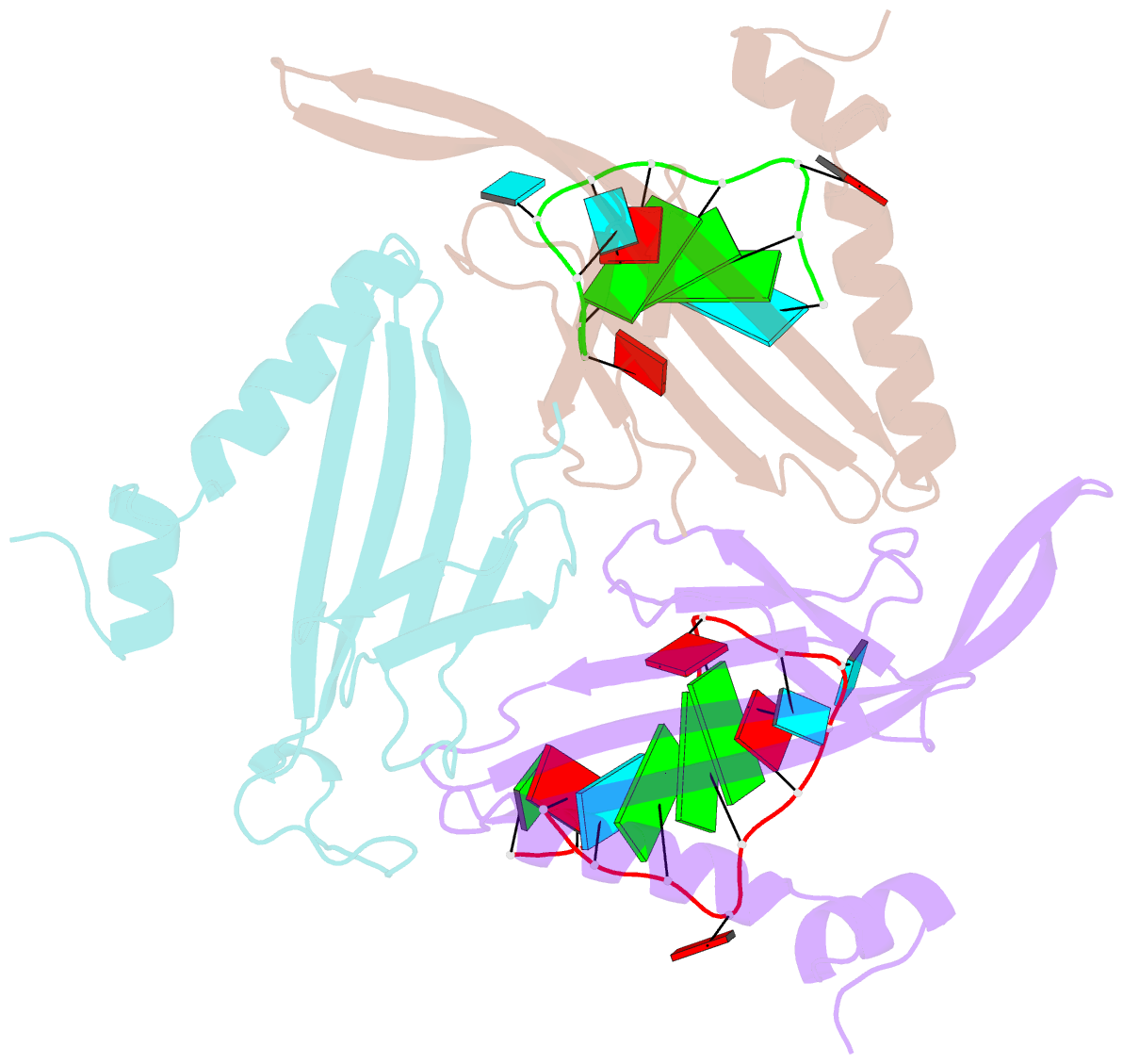
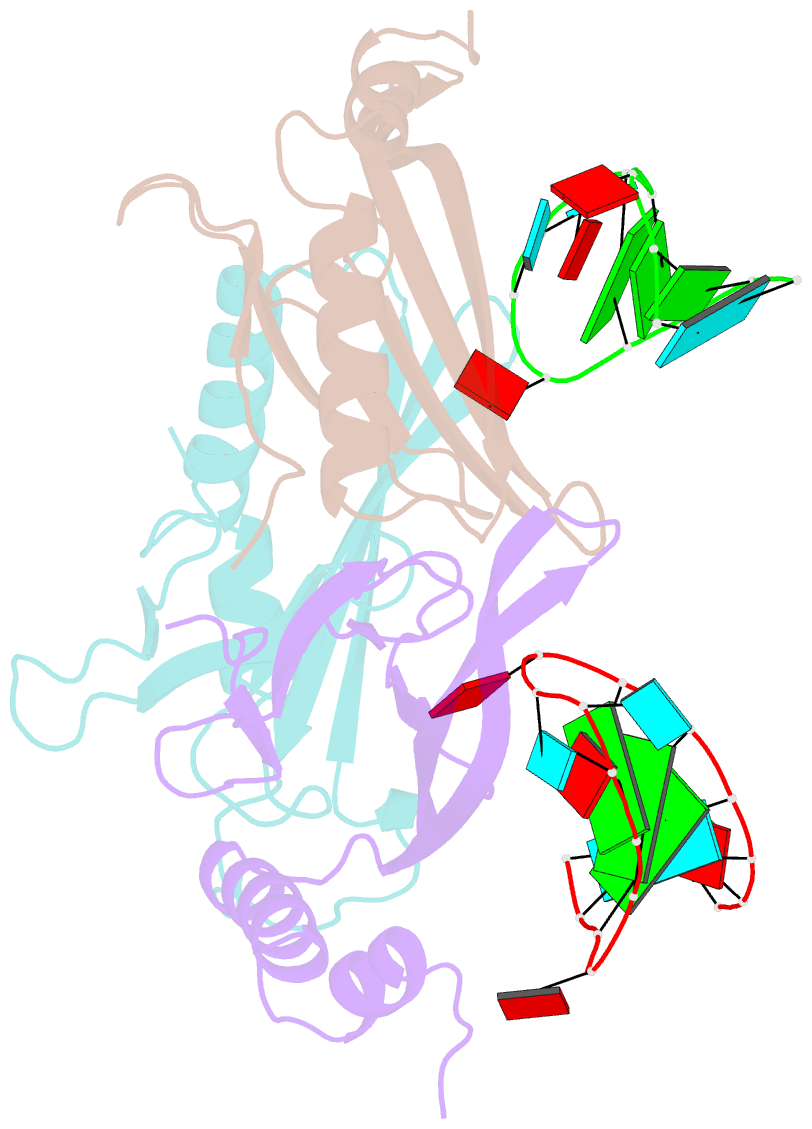
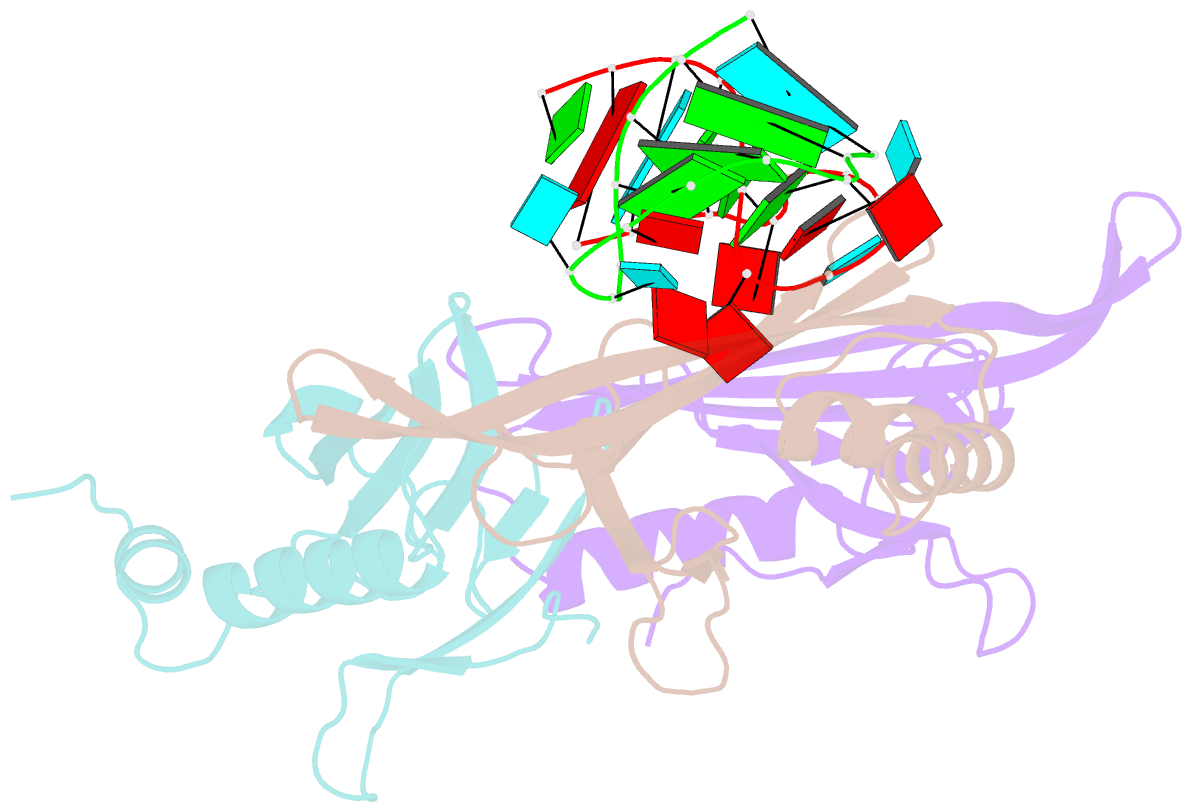
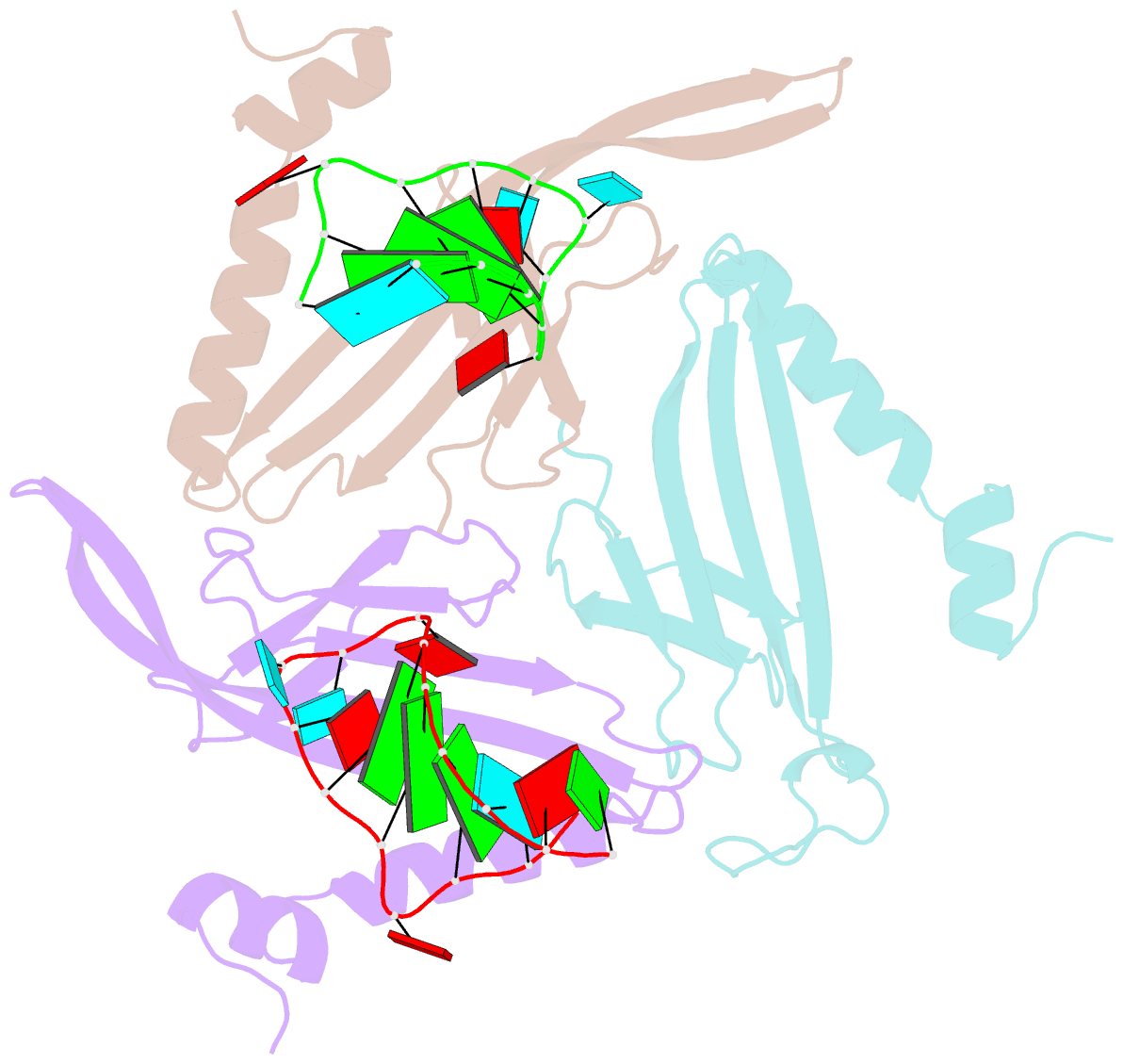
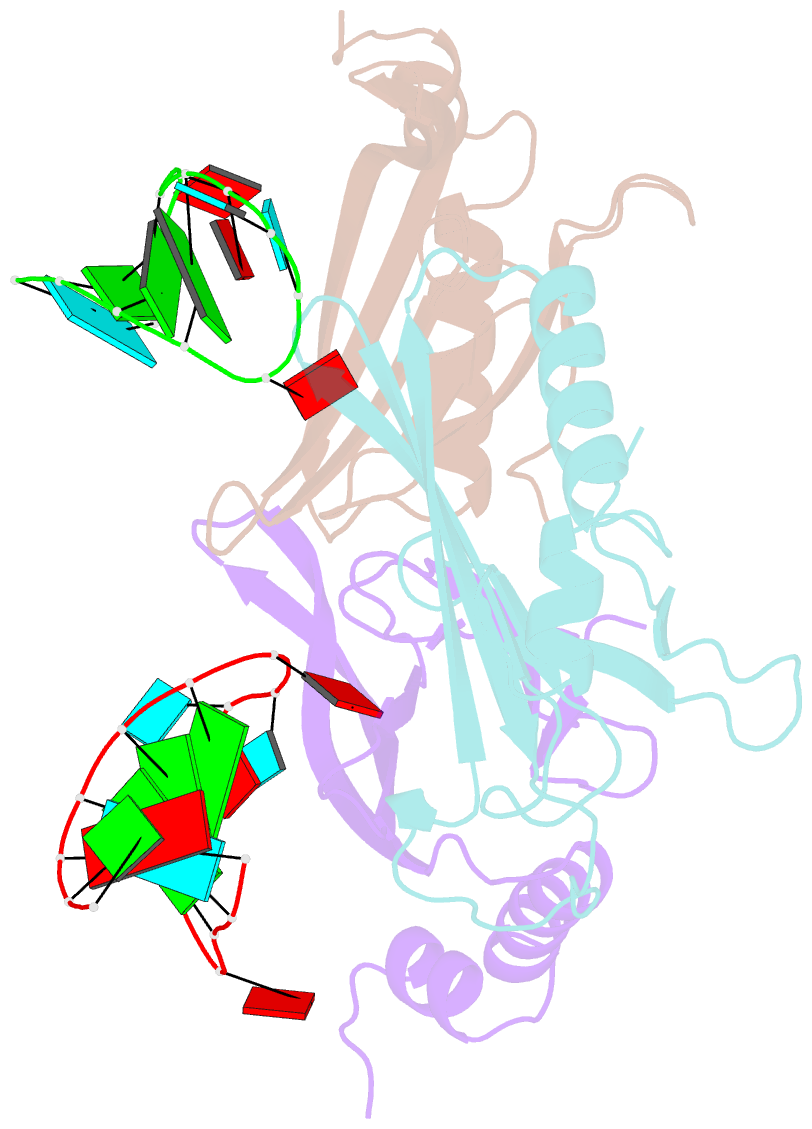
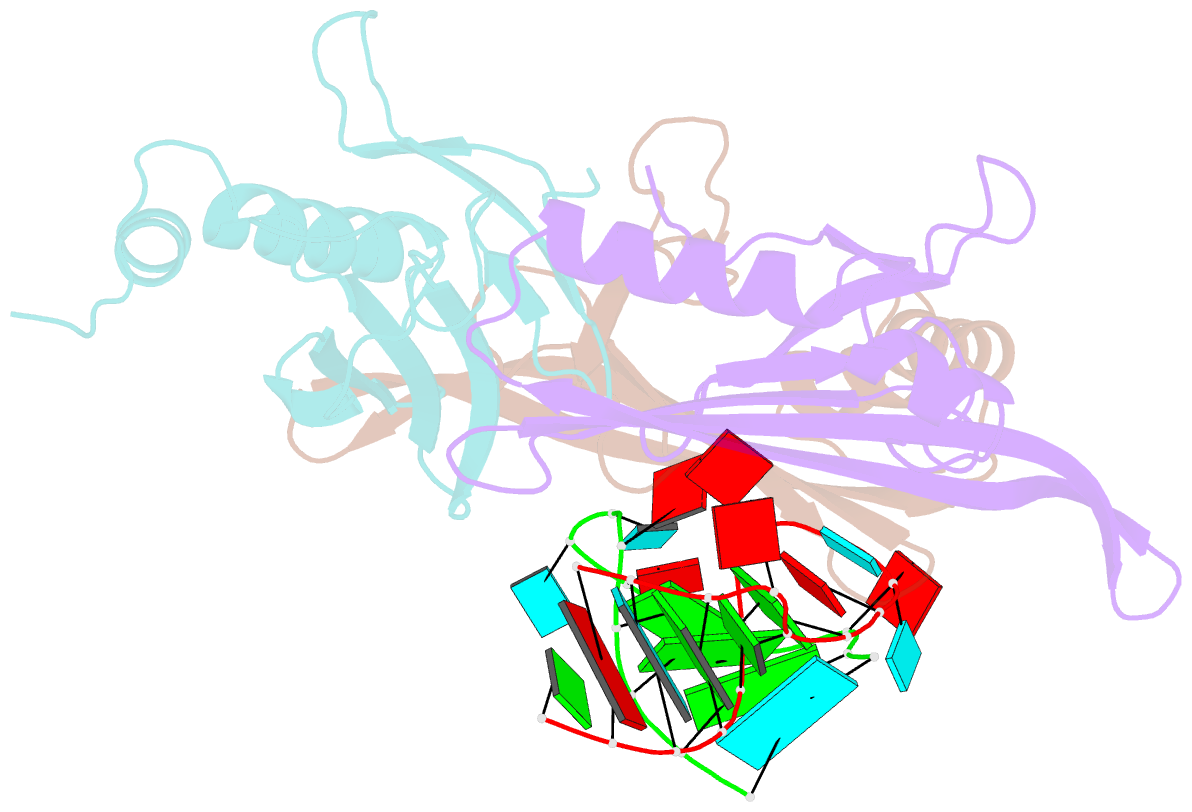
Summary information and primary citation
- PDB-id
- 1zdi; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- virus-RNA
- Method
- X-ray (2.7 Å)
- Summary
- RNA bacteriophage ms2 coat protein-RNA complex
- Reference
- Valegard K, Murray JB, Stonehouse NJ, van den Worm S, Stockley PG, Liljas L (1997): "The three-dimensional structures of two complexes between recombinant MS2 capsids and RNA operator fragments reveal sequence-specific protein-RNA interactions." J.Mol.Biol., 270, 724-738. doi: 10.1006/jmbi.1997.1144.
- Abstract
- Crystal structures of two complexes between recombinant MS2 capsids and RNA operator fragments have been determined at 2.7 A resolution. The coat protein of the RNA bacteriophage MS2 is bifunctional; it forms the icosahedral virus shell to protect the viral nucleic acid and it acts as a translational repressor by binding with high specificity to a unique site on the RNA, a single stem-loop structure, containing the initiation codon of the gene for the viral replicase. In order to determine the structure of these protein-RNA complexes, we have used chemically synthesized variants of the stem-loop fragment and soaked them into crystals of recombinant capsids. The RNA stem-loop, as bound to the protein, forms a crescent-like structure and interacts with the surface of the beta-sheet of a coat protein dimer. It makes protein contacts with seven phosphate groups on the 5' side of the stem-loop, with a pyrimidine base at position -5, which stacks onto a tyrosine, and with two exposed adenine bases, one in the loop and one at a bulge in the stem. Replacement of the wild-type uridine with a cytosine at position -5 increases the affinity of the RNA to the dimer significantly. The complex with RNA stem-loop having cytosine at this position differs from that of the wild-type complex mainly by having one extra intramolecular RNA interaction and one extra water-mediated hydrogen bond.