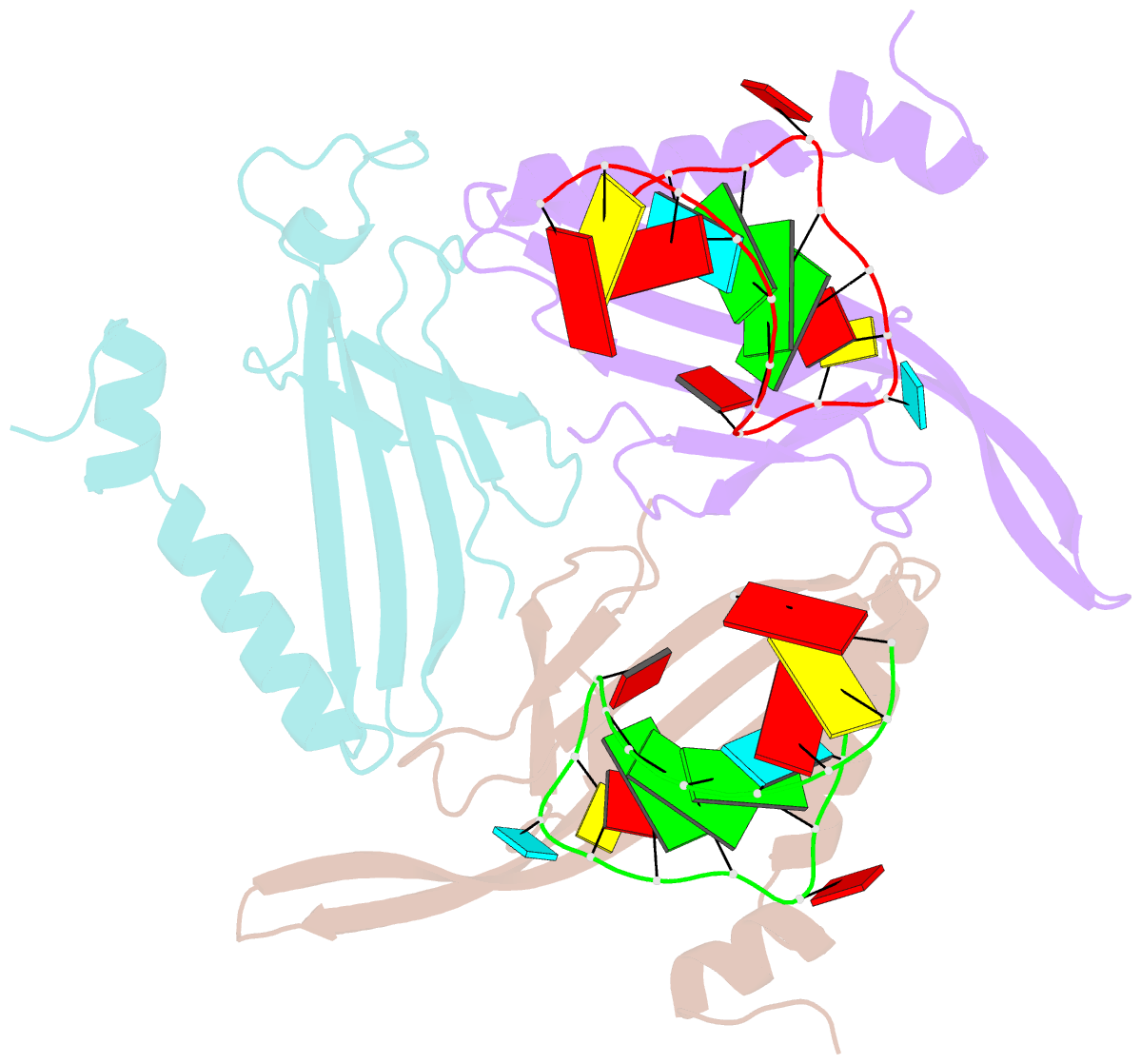
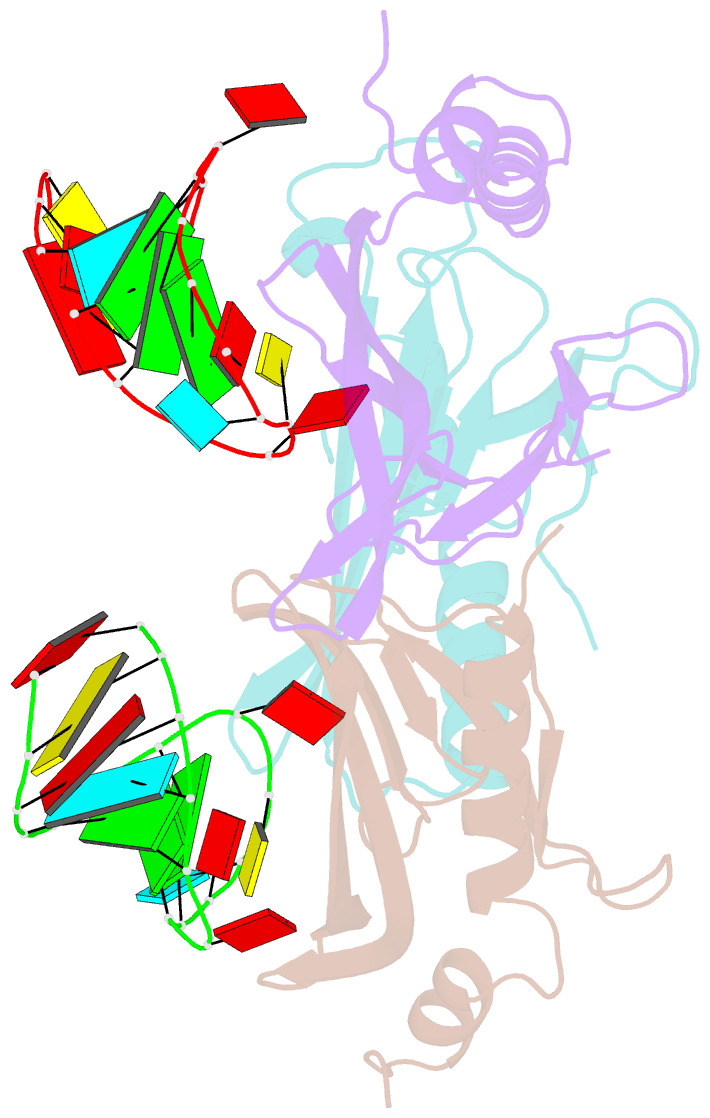
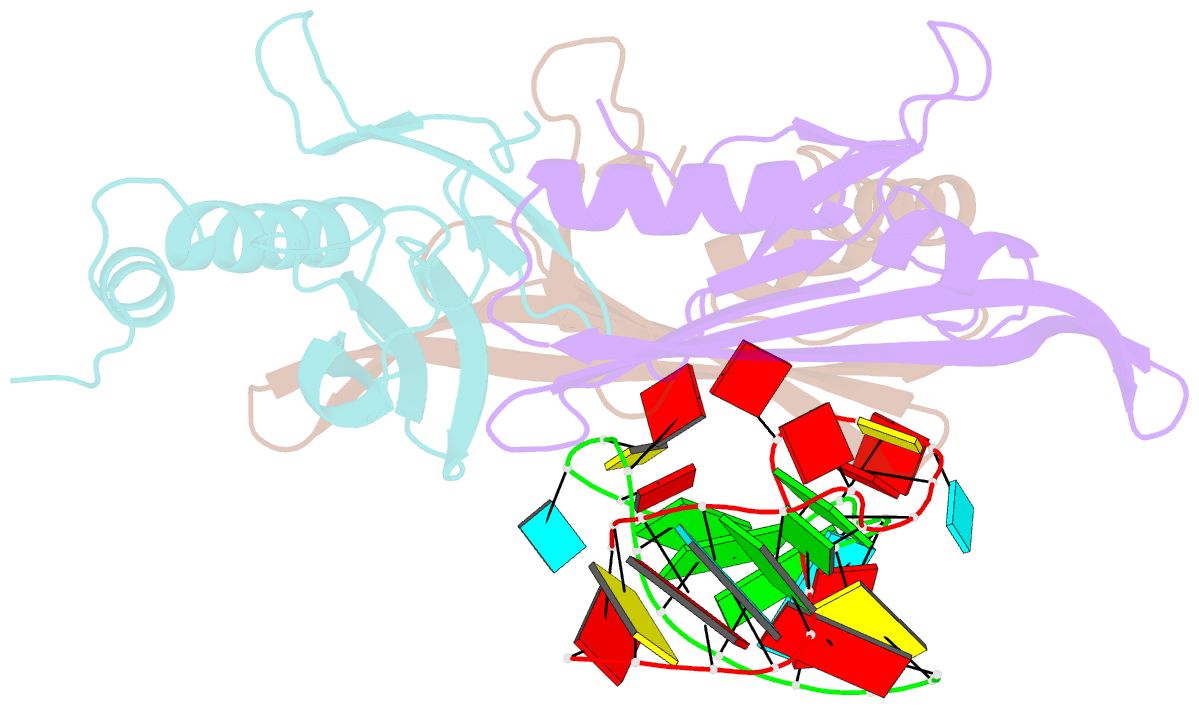
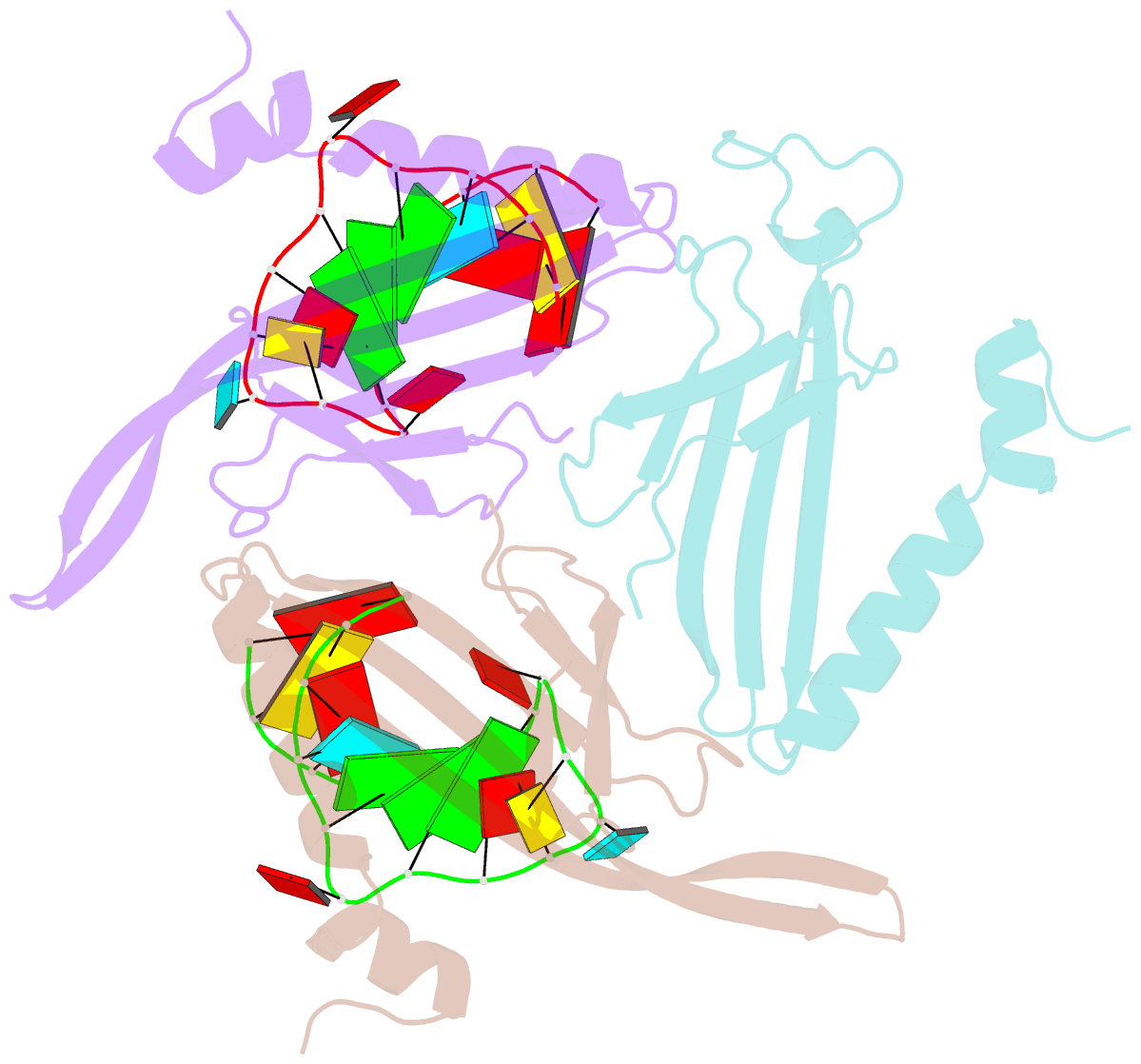
Summary information and primary citation
- PDB-id
- 1zdk; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- virus-RNA
- Method
- X-ray (2.86 Å)
- Summary
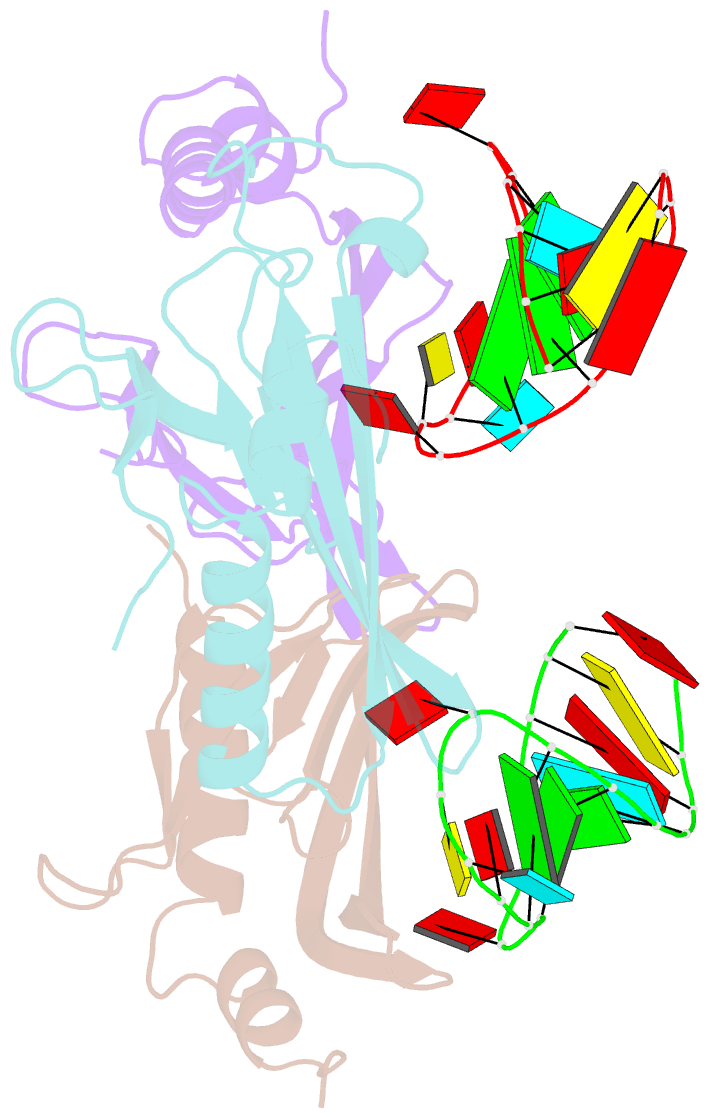
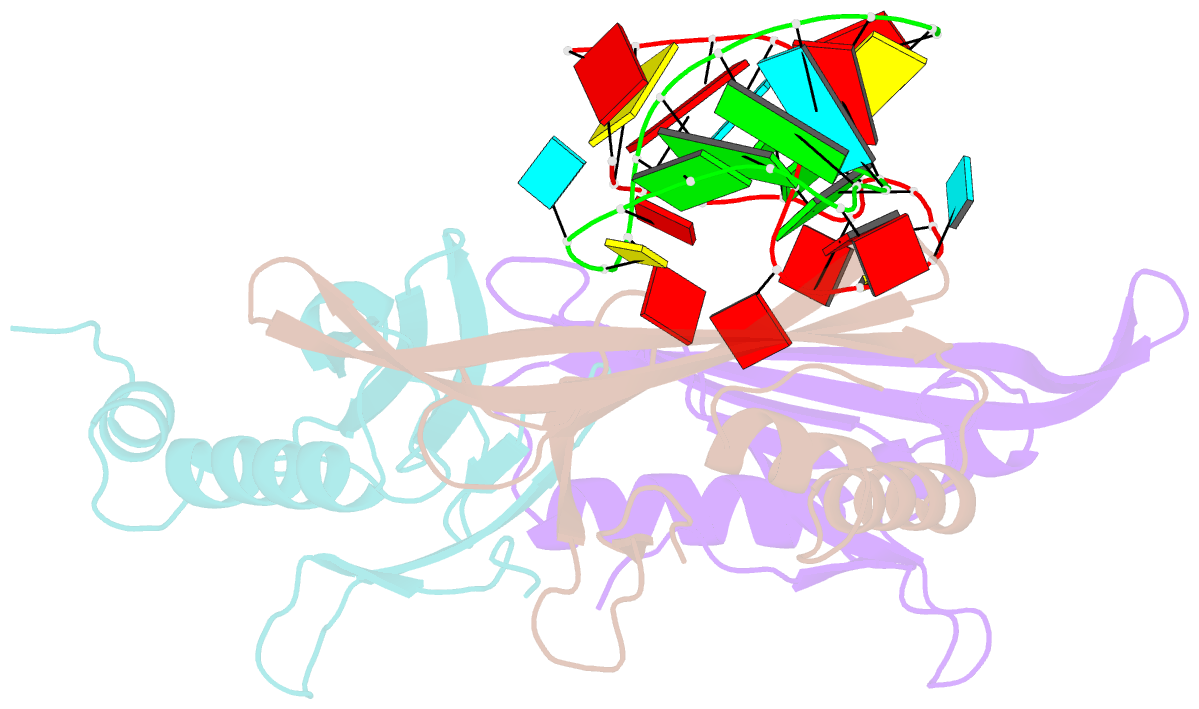
- Structure of bacteriophage coat protein-loop RNA complex
- Reference
- Grahn E, Stonehouse NJ, Murray JB, van den Worm S, Valegard K, Fridborg K, Stockley PG, Liljas L (1999): "Crystallographic studies of RNA hairpins in complexes with recombinant MS2 capsids: implications for binding requirements." RNA, 5, 131-138. doi: 10.1017/S1355838299981645.
- Abstract
- The coat protein of bacteriophage MS2 is known to bind specifically to an RNA hairpin formed within the MS2 genome. Structurally this hairpin is built up by an RNA double helix interrupted by one unpaired nucleotide and closed by a four-nucleotide loop. We have performed crystallographic studies of complexes between MS2 coat protein capsids and four RNA hairpin variants in order to evaluate the minimal requirements for tight binding to the coat protein and to obtain more information about the three-dimensional structure of these hairpins. An RNA fragment including the four loop nucleotides and a two-base-pair stem but without the unpaired nucleotide is sufficient for binding to the coat protein shell under the conditions used in this study. In contrast, an RNA fragment containing a stem with the unpaired nucleotide but missing the loop nucleotides does not bind to the protein shell.