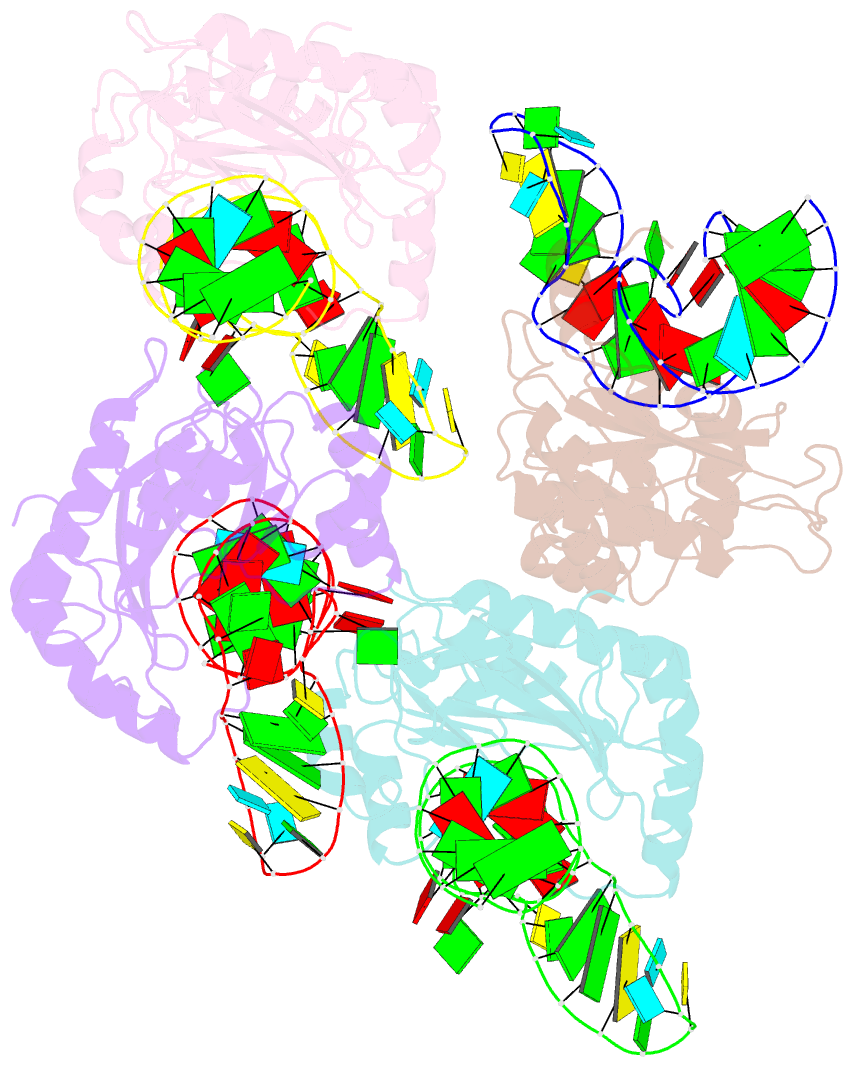
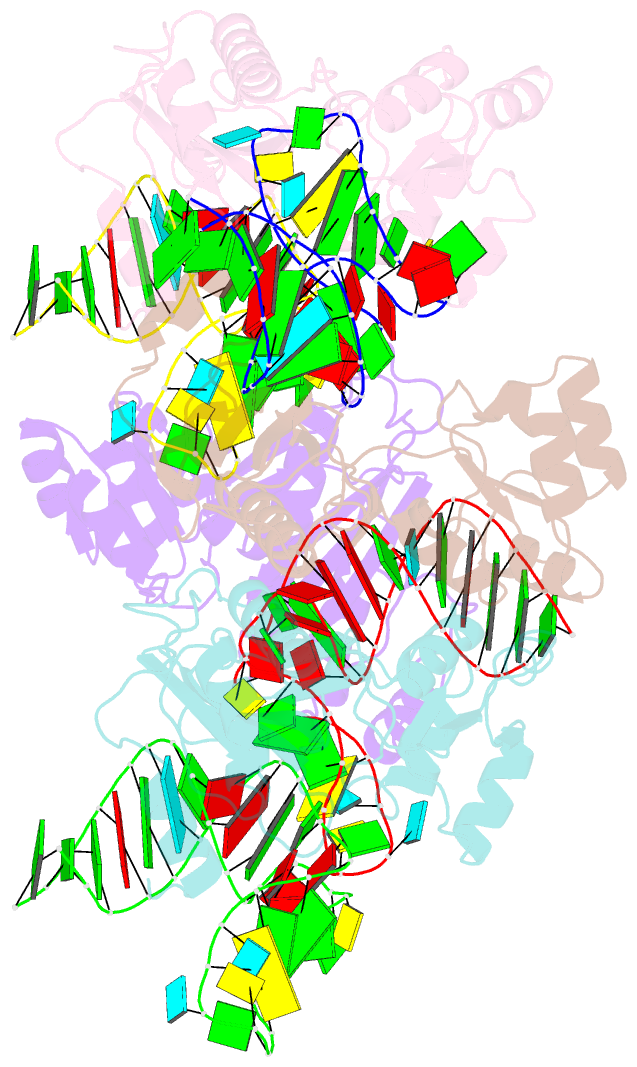
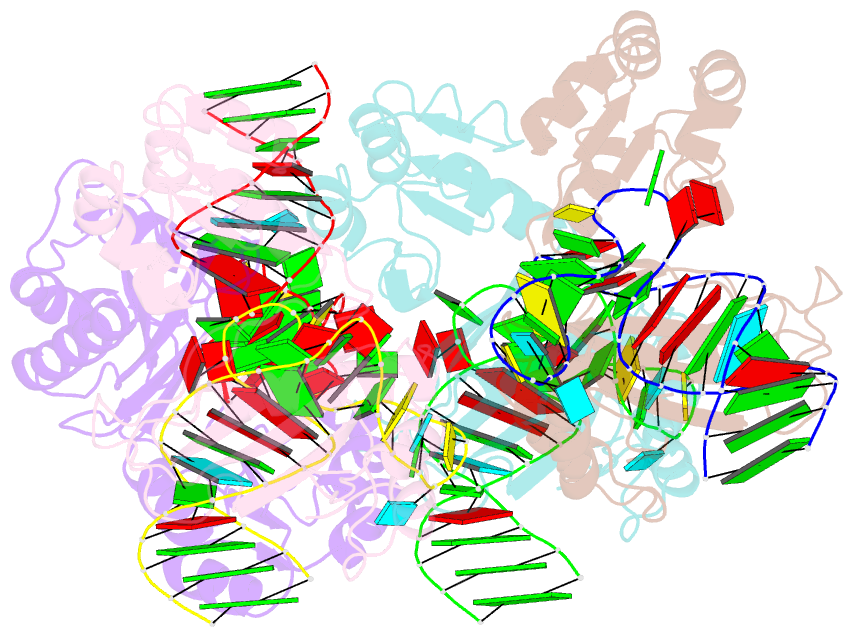
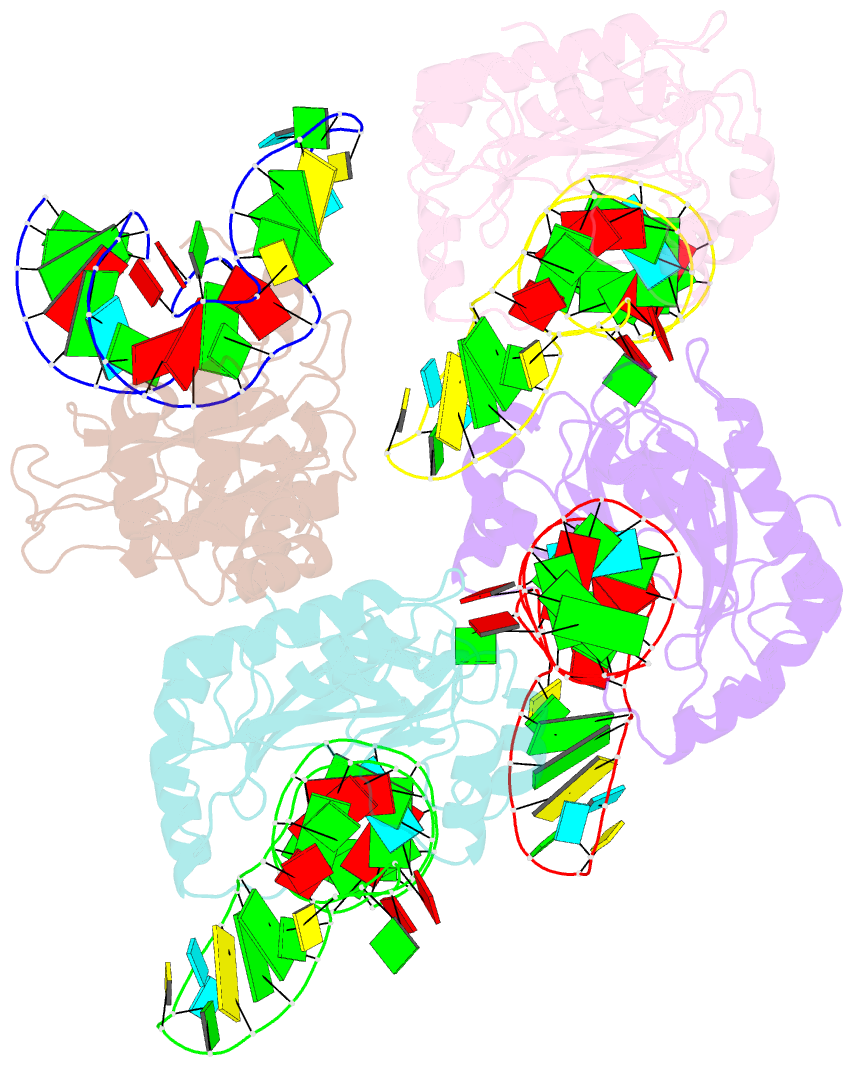
Summary information and primary citation
- PDB-id
- 1zho; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- structural protein-RNA
- Method
- X-ray (2.6 Å)
- Summary
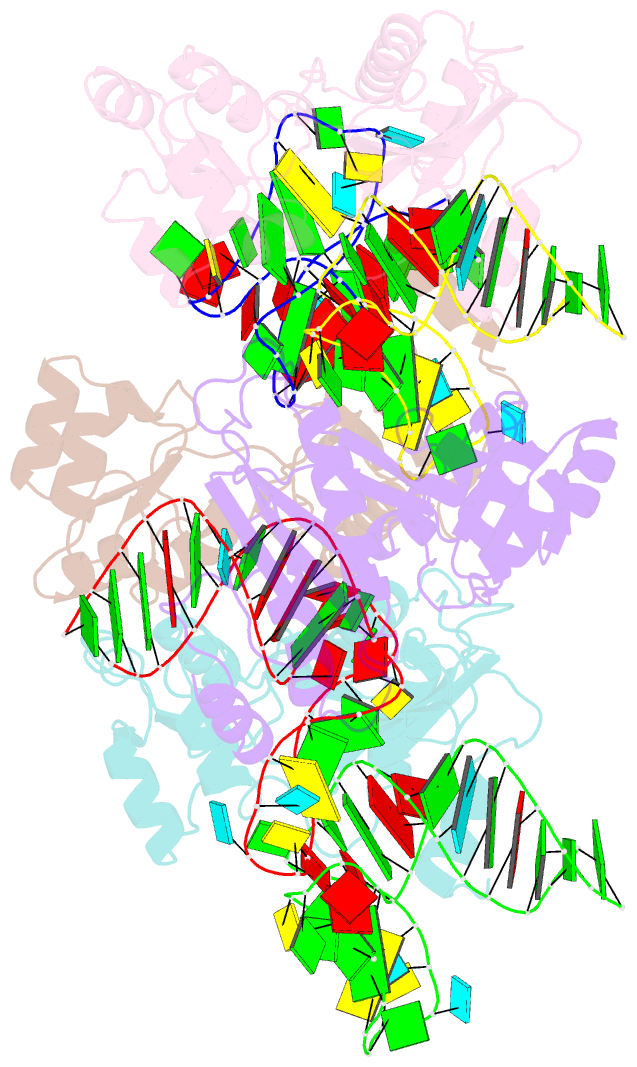
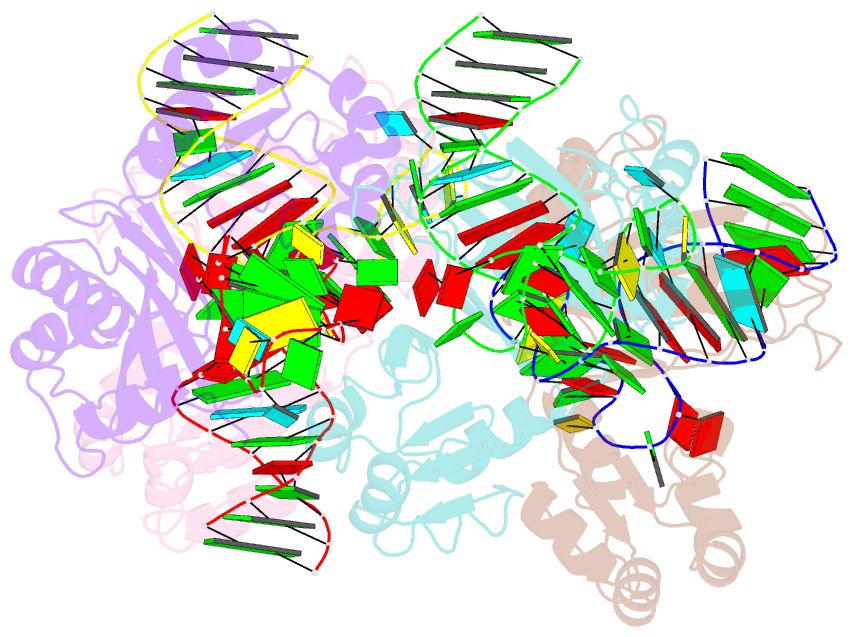
- The structure of a ribosomal protein l1 in complex with mrna
- Reference
- Nevskaya N, Tishchenko S, Volchkov S, Kljashtorny V, Nikonova E, Nikonov O, Nikulin A, Kohrer C, Piendl W, Zimmermann R, Stockley P, Garber M, Nikonov S (2006): "New insights into the interaction of ribosomal protein L1 with RNA." J.Mol.Biol., 355, 747-759. doi: 10.1016/j.jmb.2005.10.084.
- Abstract
- The RNA-binding ability of ribosomal protein L1 is of profound interest, since L1 has a dual function as a ribosomal structural protein that binds rRNA and as a translational repressor that binds its own mRNA. Here, we report the crystal structure at 2.6 A resolution of ribosomal protein L1 from the bacterium Thermus thermophilus in complex with a 38 nt fragment of L1 mRNA from Methanoccocus vannielii. The conformation of RNA-bound T.thermophilus L1 differs dramatically from that of the isolated protein. Analysis of four copies of the L1-mRNA complex in the crystal has shown that domain II of the protein does not contribute to mRNA-specific binding. A detailed comparison of the protein-RNA interactions in the L1-mRNA and L1-rRNA complexes identified amino acid residues of L1 crucial for recognition of its specific targets on the both RNAs. Incorporation of the structure of bacterial L1 into a model of the Escherichia coli ribosome revealed two additional contact regions for L1 on the 23S rRNA that were not identified in previous ribosome models.