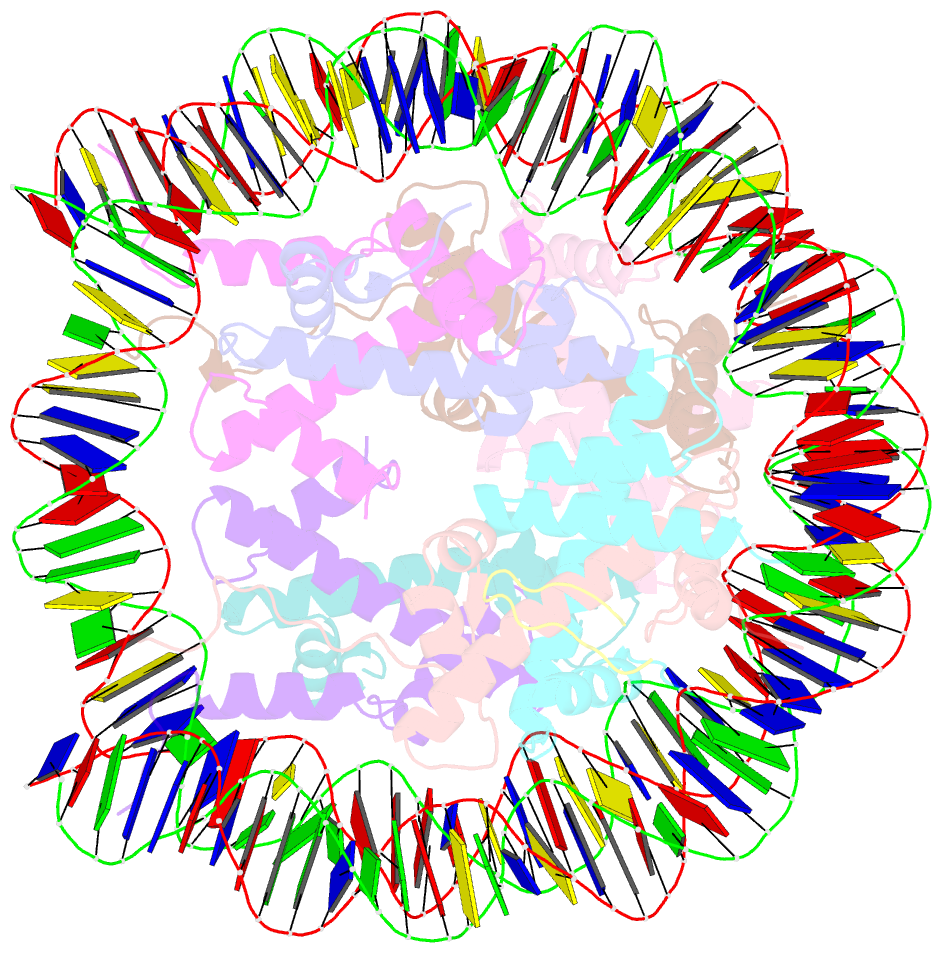
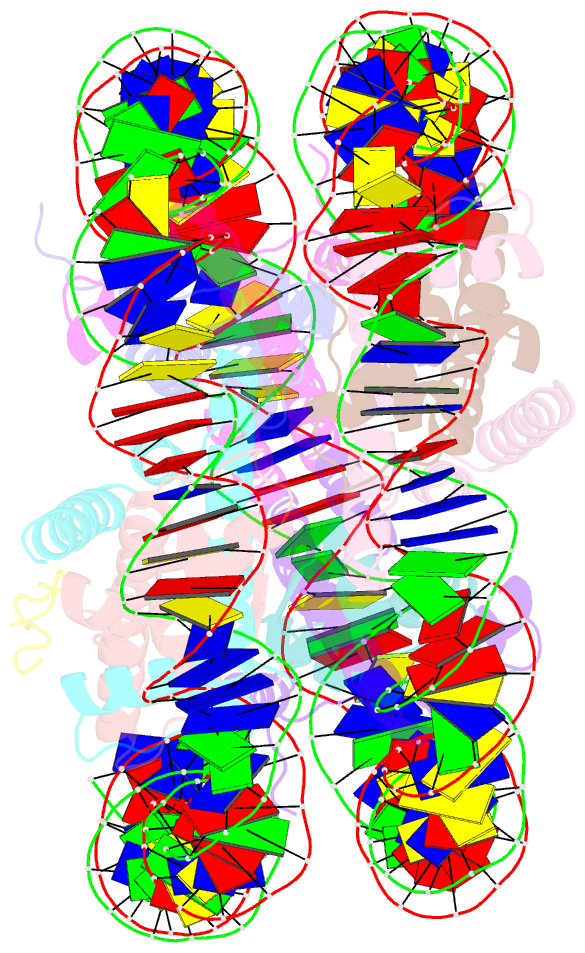
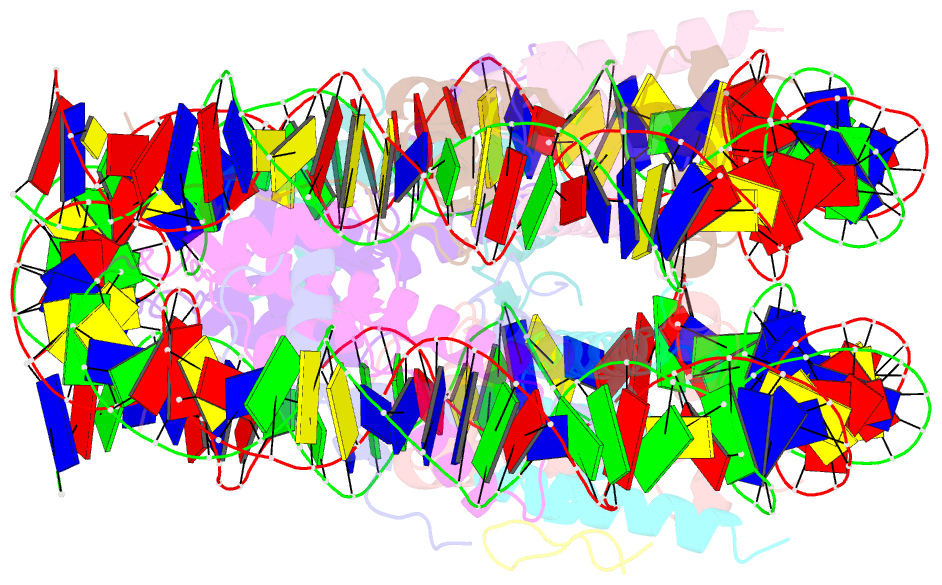
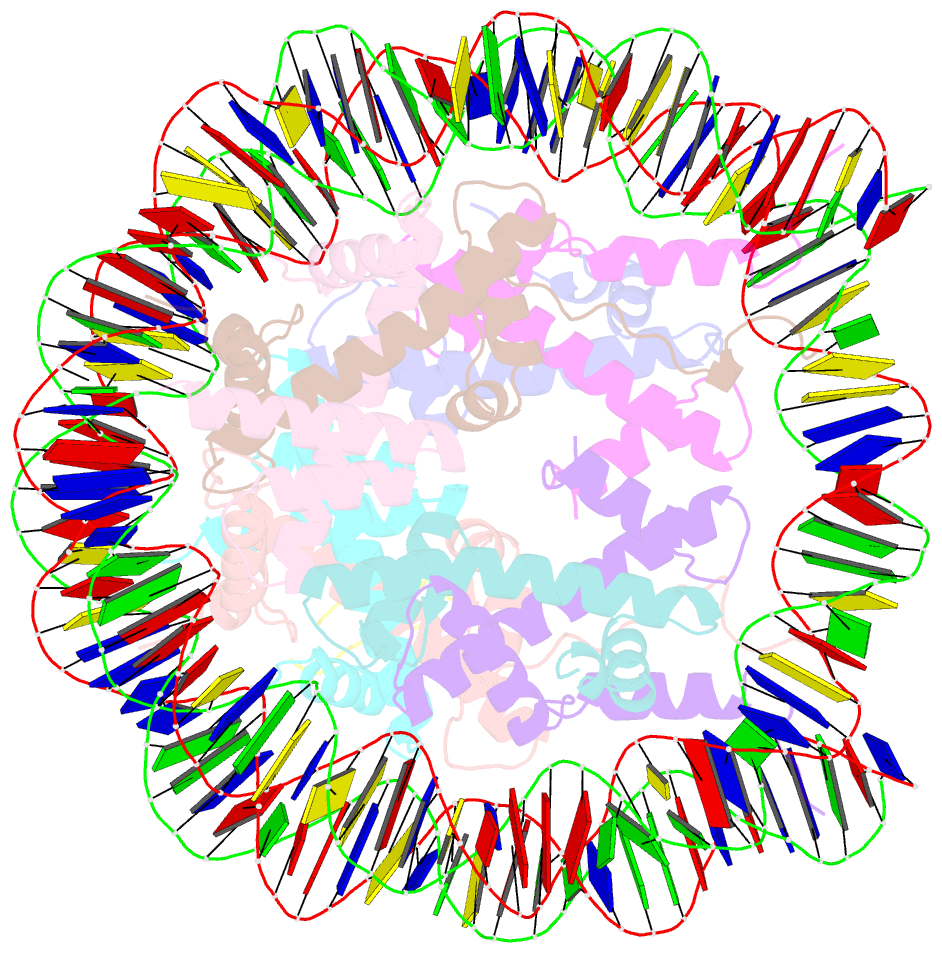
Summary information and primary citation
- PDB-id
- 1zla; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- structural protein-DNA
- Method
- X-ray (2.9 Å)
- Summary
- X-ray structure of a kaposi's sarcoma herpesvirus lana peptide bound to the nucleosomal core
- Reference
- Barbera AJ, Chodaparambil JV, Kelley-Clarke B, Joukov V, Walter JC, Luger K, Kaye KM (2006): "The nucleosomal surface as a docking station for Kaposi's sarcoma herpesvirus LANA." Science, 311, 856-861. doi: 10.1126/science.1120541.
- Abstract
- Kaposi's sarcoma-associated herpesvirus (KSHV) latency-associated nuclear antigen (LANA) mediates viral genome attachment to mitotic chromosomes. We find that N-terminal LANA docks onto chromosomes by binding nucleosomes through the folded region of histones H2A-H2B. The same LANA residues were required for both H2A-H2B binding and chromosome association. Further, LANA did not bind Xenopus sperm chromatin, which is deficient in H2A-H2B; chromatin binding was rescued after assembly of nucleosomes containing H2A-H2B. We also describe the 2.9-angstrom crystal structure of a nucleosome complexed with the first 23 LANA amino acids. The LANA peptide forms a hairpin that interacts exclusively with an acidic H2A-H2B region that is implicated in the formation of higher order chromatin structure. Our findings present a paradigm for how nucleosomes may serve as binding platforms for viral and cellular proteins and reveal a previously unknown mechanism for KSHV latency.