Summary information and primary citation
- PDB-id
- 1zq3; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transcription-DNA
- Method
- NMR
- Summary
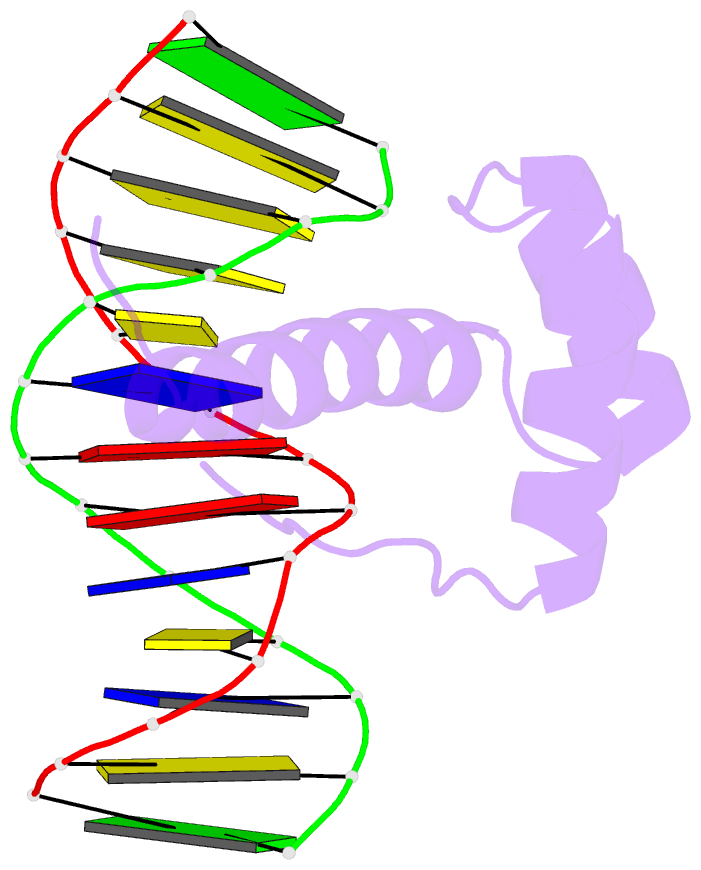
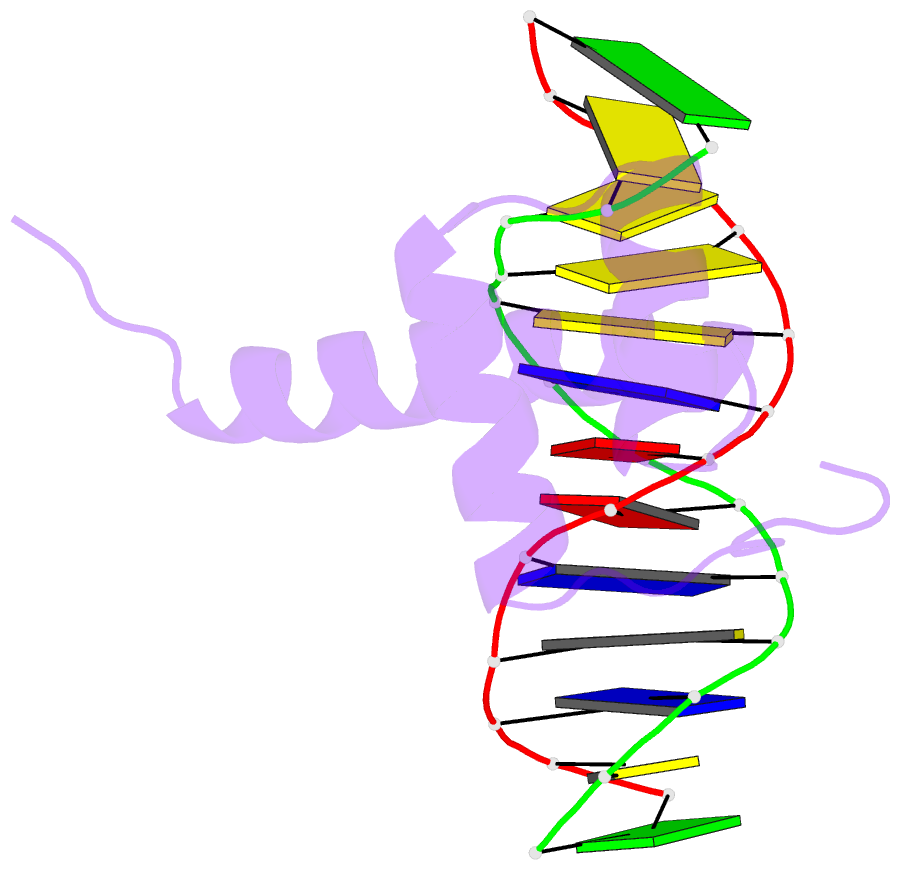
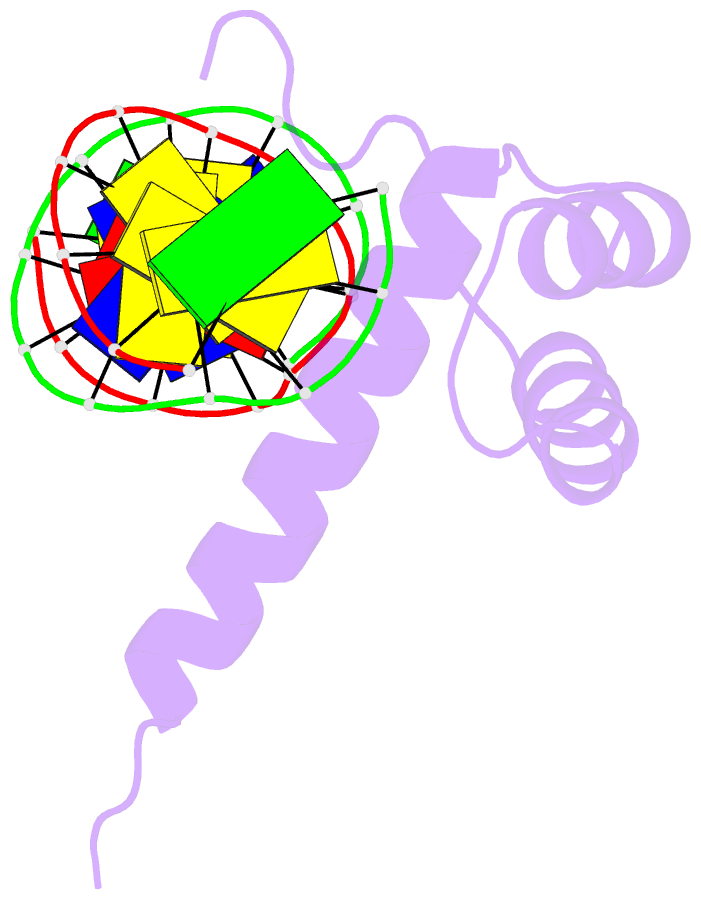
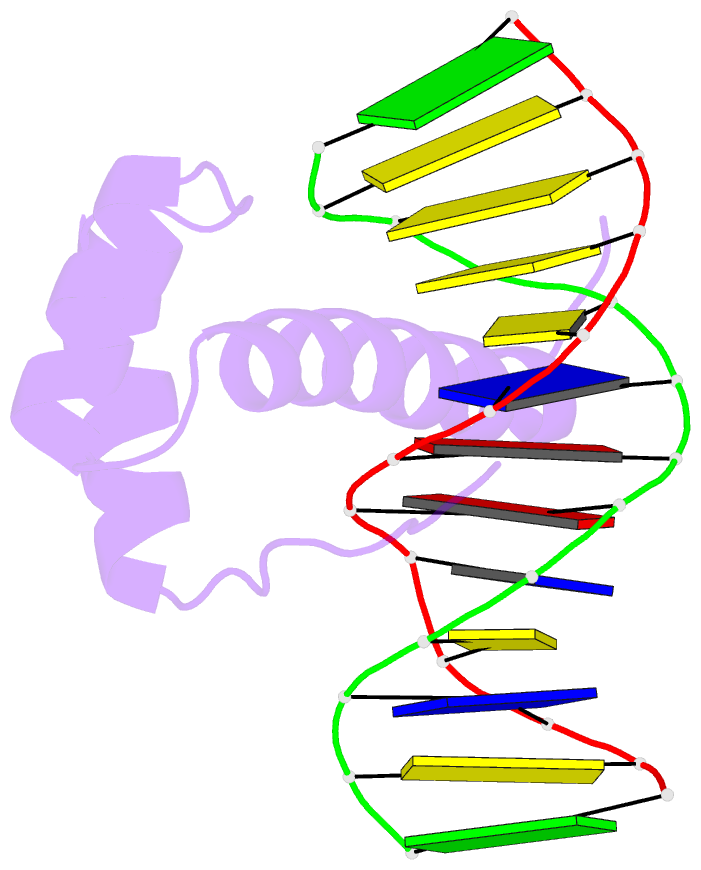
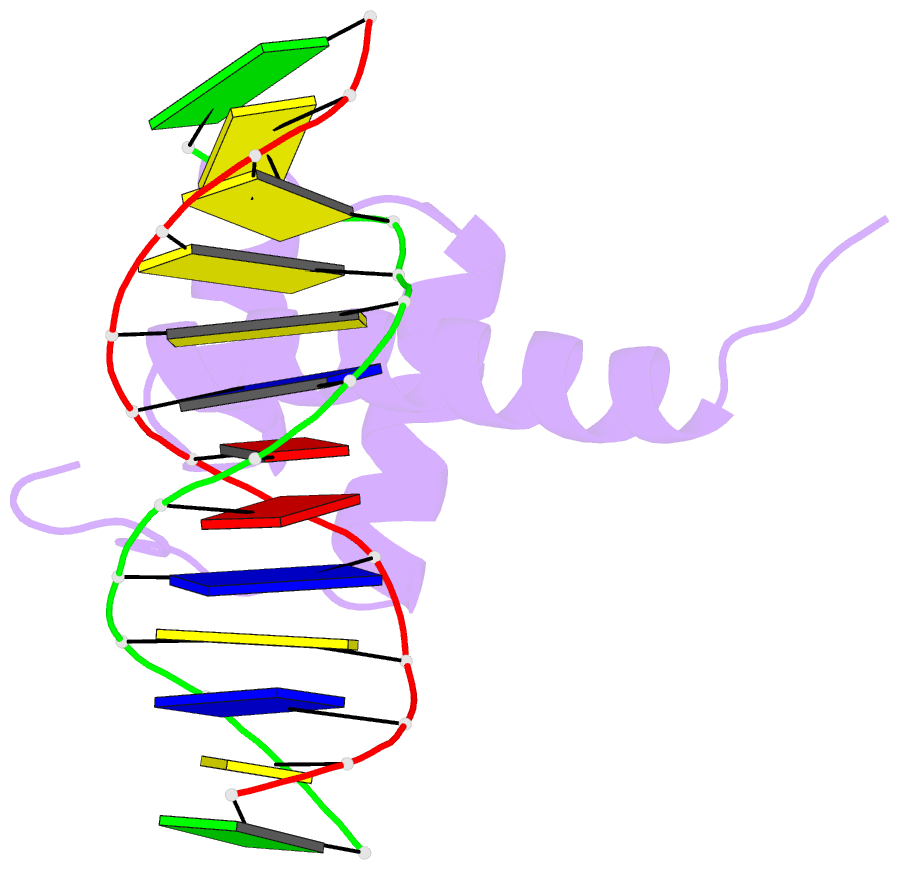
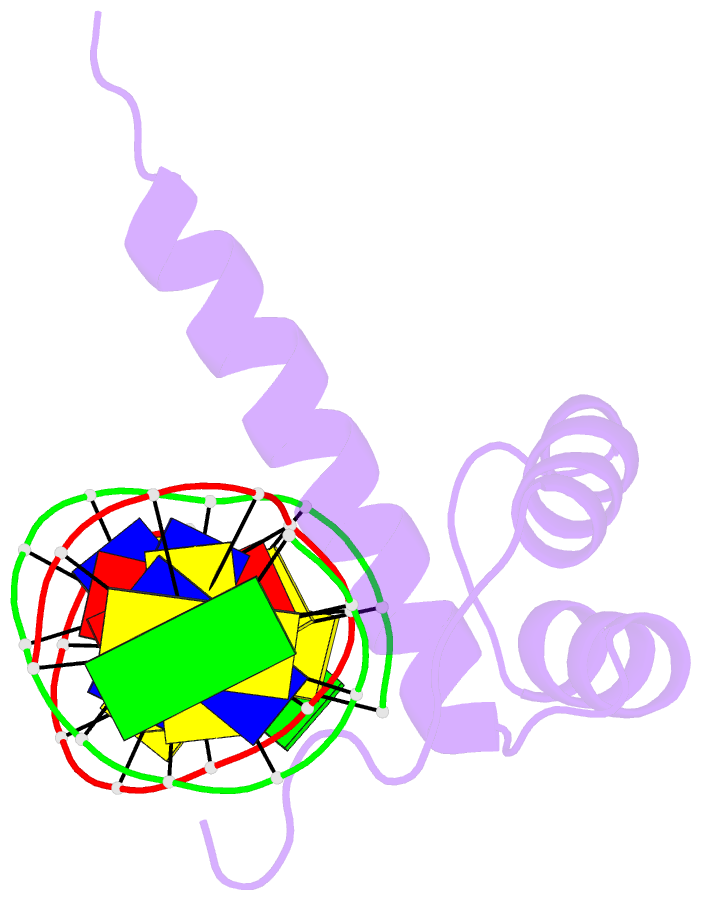
- NMR solution structure of the bicoid homeodomain bound to the consensus DNA binding site taatcc
- Reference
- Baird-Titus JM, Clark-Baldwin K, Dave V, Caperelli CA, Ma J, Rance M (2006): "The solution structure of the native K50 Bicoid homeodomain bound to the consensus TAATCC DNA-binding site." J.Mol.Biol., 356, 1137-1151. doi: 10.1016/j.jmb.2005.12.007.
- Abstract
- The solution structure of the homeodomain of the Drosophila morphogenic protein Bicoid (Bcd) complexed with a TAATCC DNA site is described. Bicoid is the only known protein that uses a homeodomain to regulate translation, as well as transcription, by binding to both RNA and DNA during early Drosophila development; in addition, the Bcd homeodomain can recognize an array of different DNA sites. The dual functionality and broad recognition capabilities signify that the Bcd homeodomain may possess unique structural/dynamic properties. Bicoid is the founding member of the K50 class of homeodomain proteins, containing a lysine residue at the critical 50th position (K50) of the homeodomain sequence, a residue required for DNA and RNA recognition; Bcd also has an arginine residue at the 54th position (R54), which is essential for RNA recognition. Bcd is the only known homeodomain with the K50/R54 combination of residues. The Bcd structure indicates that this homeodomain conforms to the conserved topology of the homeodomain motif, but exhibits a significant variation from other homeodomain structures at the end of helix 1. A key result is the observation that the side-chains of the DNA-contacting residues K50, N51 and R54 all show strong signs of flexibility in the protein-DNA interface. This finding is supportive of the adaptive-recognition theory of protein-DNA interactions.