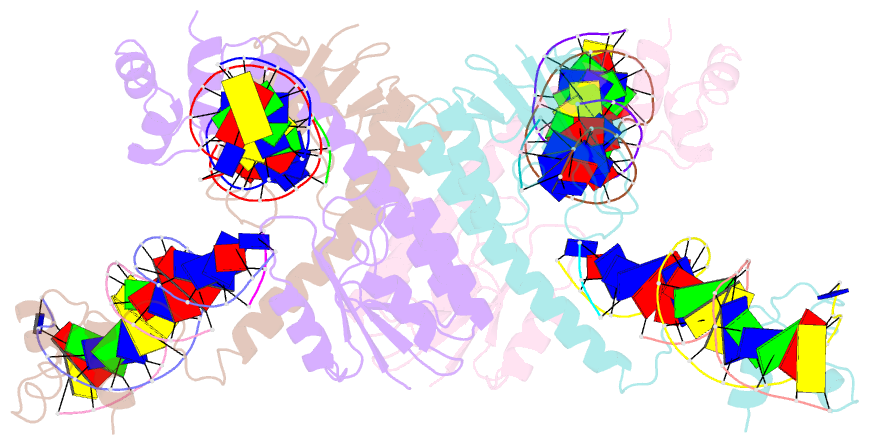
Summary information and primary citation
- PDB-id
- 1zr4; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- recombination-DNA
- Method
- X-ray (3.4 Å)
- Summary
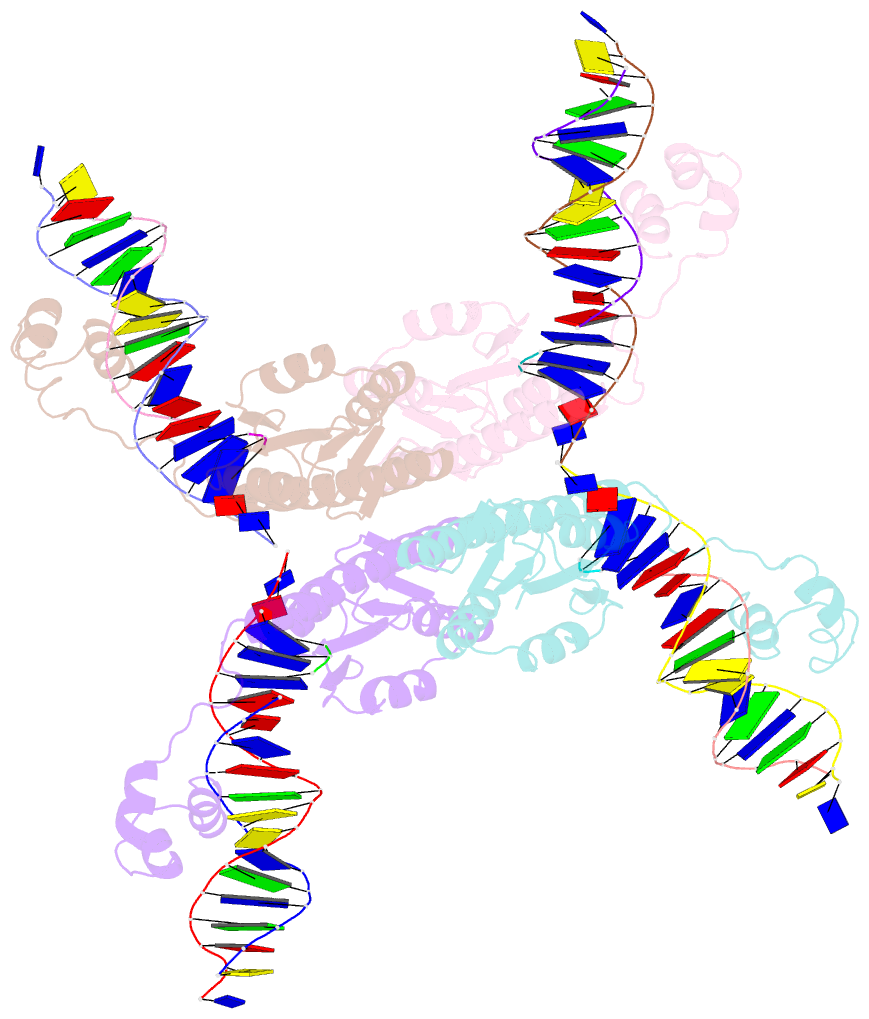
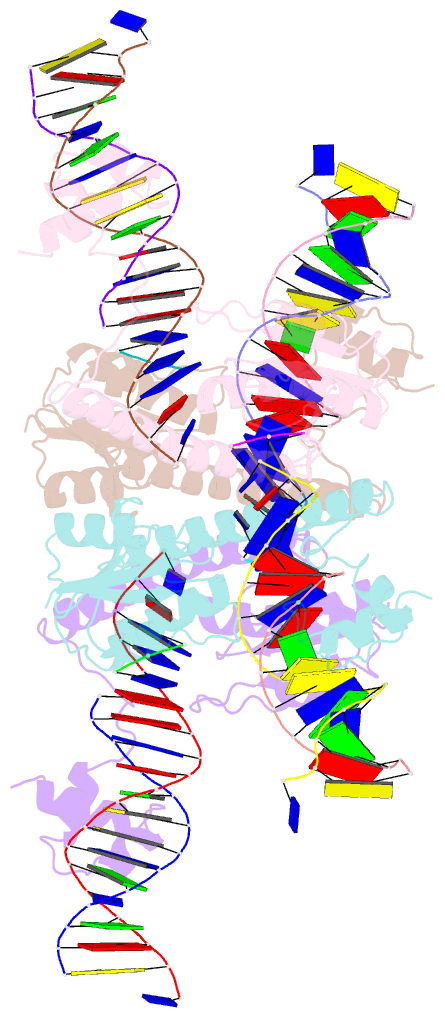
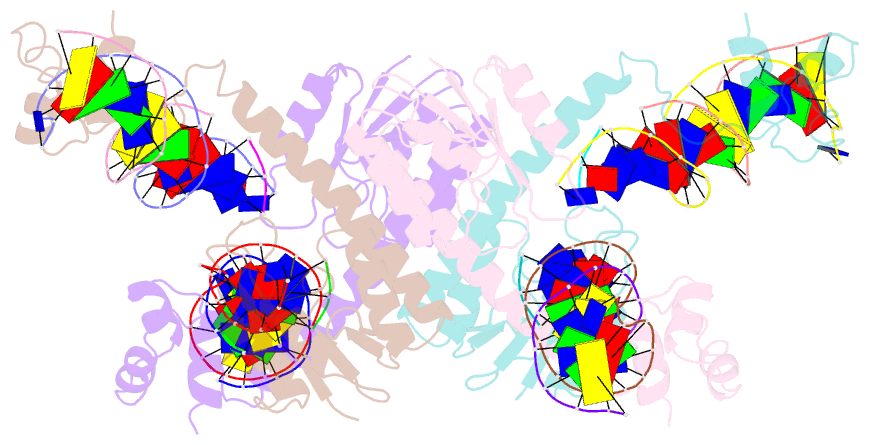
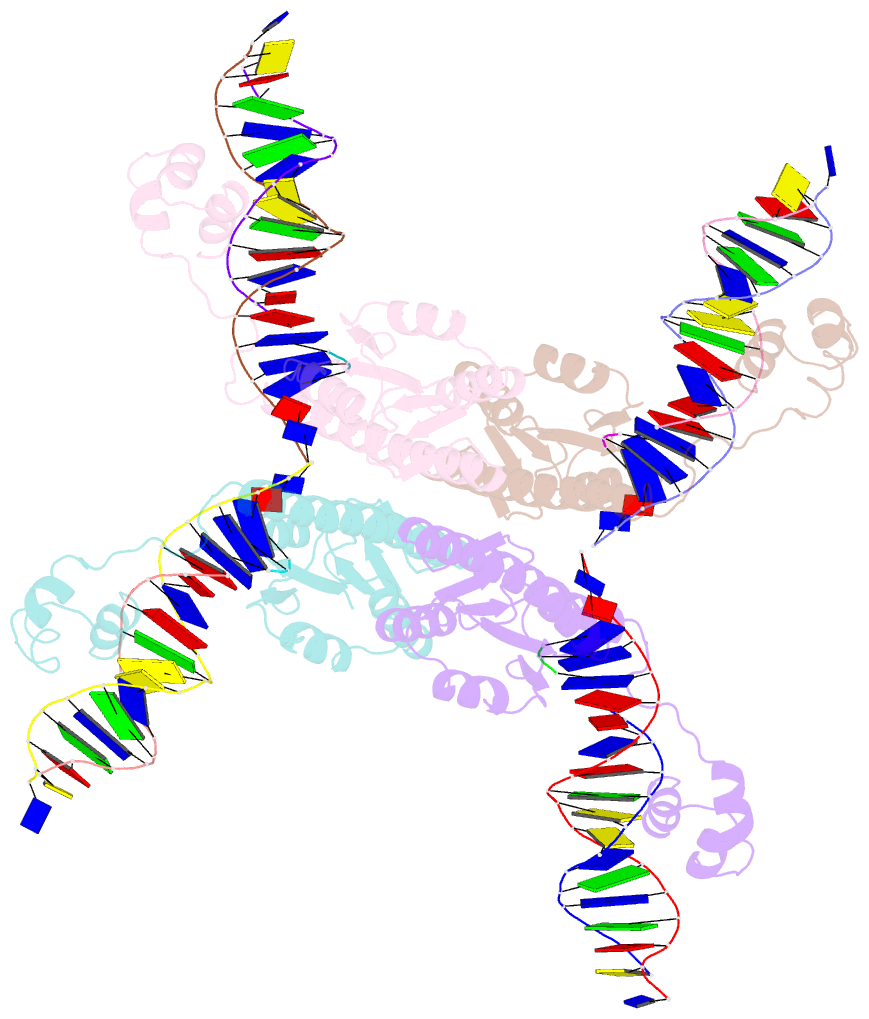
- Structure of a synaptic gamma-delta resolvase tetramer covalently linked to two cleaved dnas
- Reference
- Li W, Kamtekar S, Xiong Y, Sarkis GJ, Grindley ND, Steitz TA (2005): "Structure of a synaptic gamma delta resolvase tetramer covalently linked to two cleaved DNAs." Science, 309, 1210-1215. doi: 10.1126/science.1112064.
- Abstract
- The structure of a synaptic intermediate of the site-specific recombinase gammadelta resolvase covalently linked through Ser10 to two cleaved duplex DNAs has been determined at 3.4 angstrom resolution. This resolvase, activated for recombination by mutations, forms a tetramer whose structure is substantially changed from that of a presynaptic complex between dimeric resolvase and the cleavage site DNA. Because the two cleaved DNA duplexes that are to be recombined lie on opposite sides of the core tetramer, large movements of both protein and DNA are required to achieve strand exchange. The two dimers linked to the DNAs that are to be recombined are held together by a flat interface. This may allow a 180 degrees rotation of one dimer relative to the other in order to reposition the DNA duplexes for strand exchange.