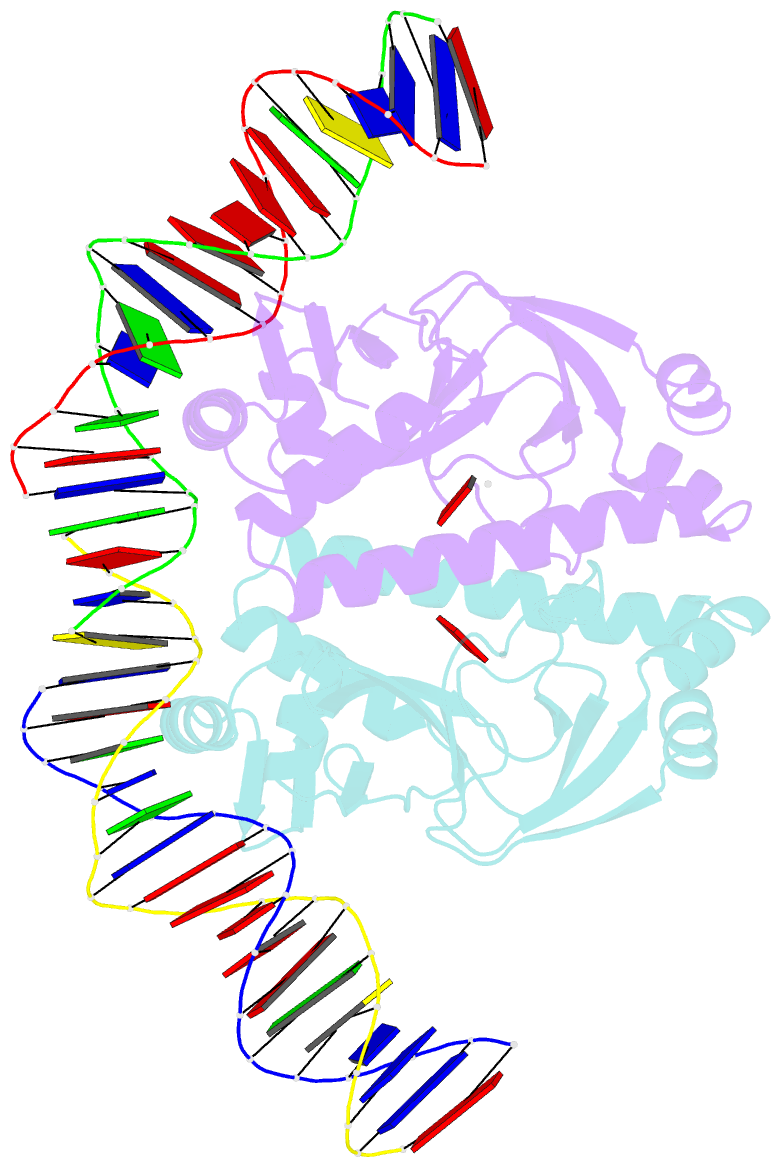
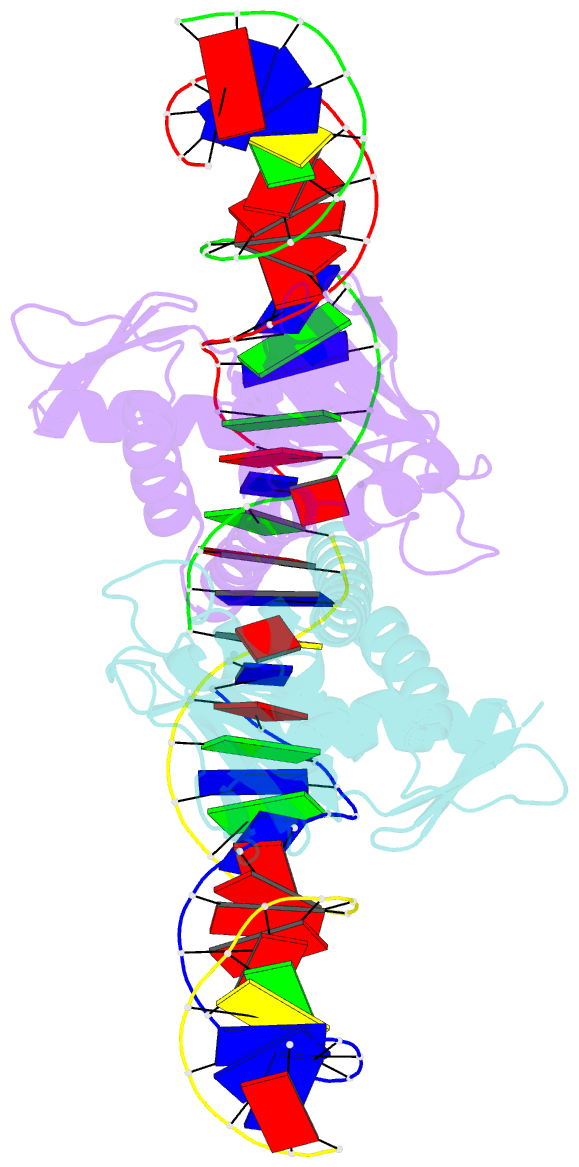
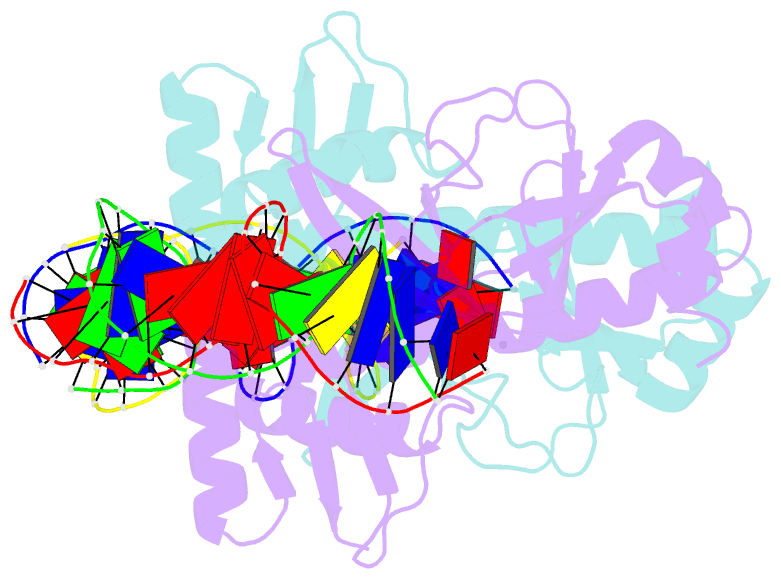
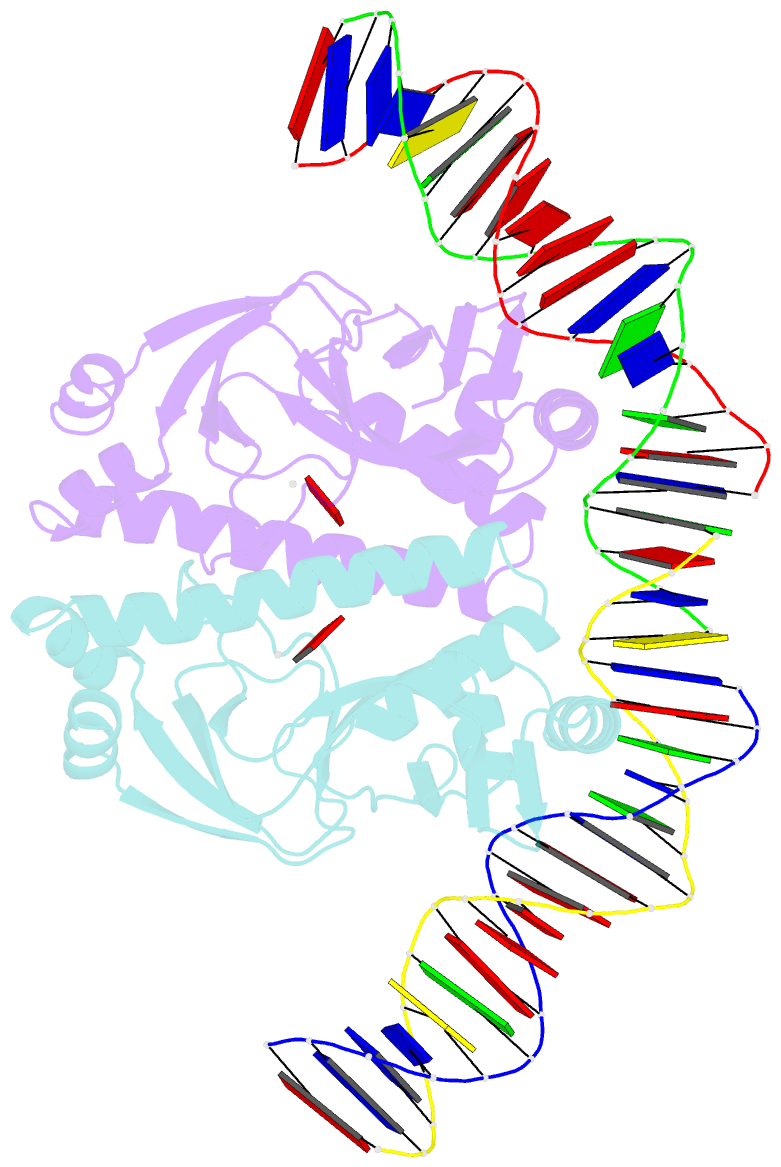
Summary information and primary citation
- PDB-id
- 1zrc; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- gene regulation-DNA
- Method
- X-ray (2.8 Å)
- Summary
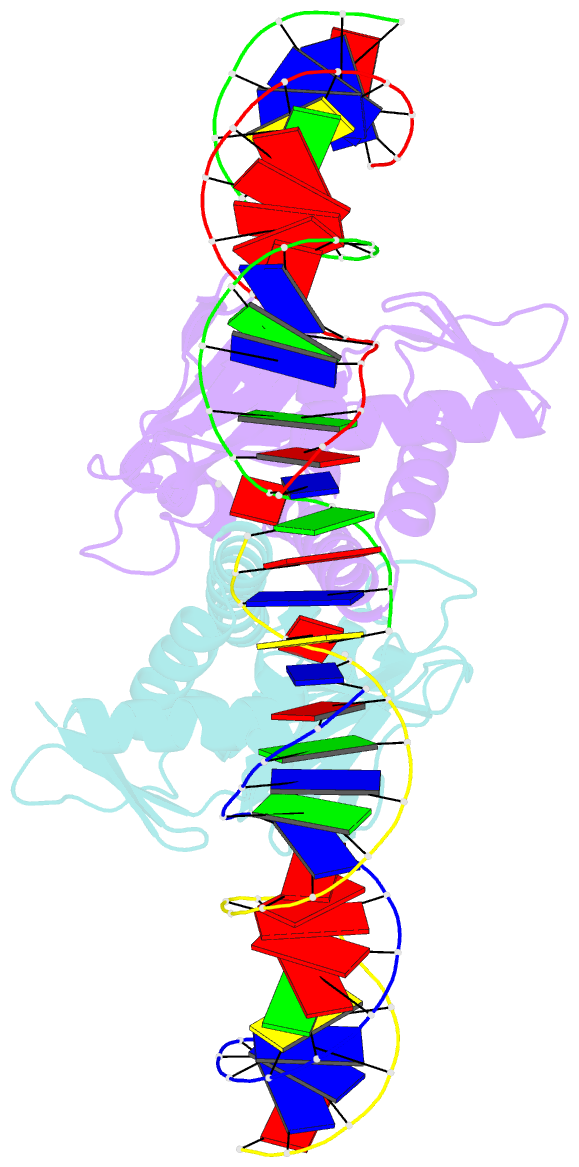
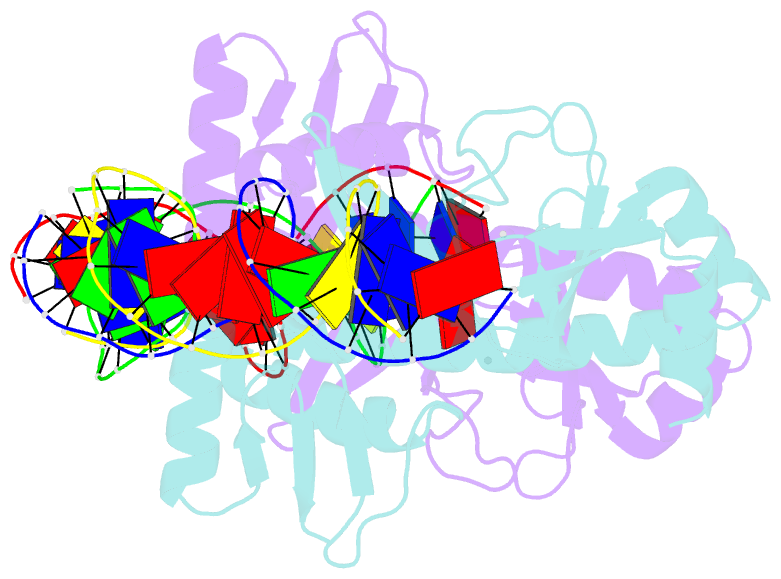
- 4 crystal structures of cap-DNA with all base-pair substitutions at position 6, cap-icap38 DNA
- Reference
- Napoli AA, Lawson CL, Ebright RH, Berman HM (2006): "Indirect readout of DNA sequence at the primary-kink site in the CAP-DNA complex: recognition of pyrimidine-purine and purine-purine steps." J.Mol.Biol., 357, 173-183. doi: 10.1016/j.jmb.2005.12.051.
- Abstract
- The catabolite activator protein (CAP) bends DNA in the CAP-DNA complex, typically introducing a sharp DNA kink, with a roll angle of approximately 40 degrees and a twist angle of approximately 20 degrees, between positions 6 and 7 of the DNA half-site, 5'-A1A2A3T4G5T6G7A8T9C10T11 -3' ("primary kink"). In previous work, we showed that CAP recognizes the nucleotide immediately 5' to the primary-kink site, T6, through an "indirect-readout" mechanism involving sequence effects on energetics of primary-kink formation. Here, to understand further this example of indirect readout, we have determined crystal structures of CAP-DNA complexes containing each possible nucleotide at position 6. The structures show that CAP can introduce a DNA kink at the primary-kink site with any nucleotide at position 6. The DNA kink is sharp with the consensus pyrimidine-purine step T6G7 and the non-consensus pyrimidine-purine step C6G7 (roll angles of approximately 42 degrees, twist angles of approximately 16 degrees ), but is much less sharp with the non-consensus purine-purine steps A6G7 and G6G7 (roll angles of approximately 20 degrees, twist angles of approximately 17 degrees). We infer that CAP discriminates between consensus and non-consensus pyrimidine-purine steps at positions 6-7 solely based on differences in the energetics of DNA deformation, but that CAP discriminates between the consensus pyrimidine-purine step and non-consensus purine-purine steps at positions 6-7 both based on differences in the energetics of DNA deformation and based on qualitative differences in DNA deformation. The structures further show that CAP can achieve a similar, approximately 46 degrees per DNA half-site, overall DNA bend through a sharp DNA kink, a less sharp DNA kink, or a smooth DNA bend. Analysis of these and other crystal structures of CAP-DNA complexes indicates that there is a large, approximately 28 degrees per DNA half-site, out-of-plane component of CAP-induced DNA bending in structures not constrained by end-to-end DNA lattice interactions and that lattice contacts involving CAP tend to involve residues in or near biologically functional surfaces.