Summary information and primary citation
- PDB-id
- 2a6o; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transcription-DNA
- Method
- X-ray (2.6 Å)
- Summary
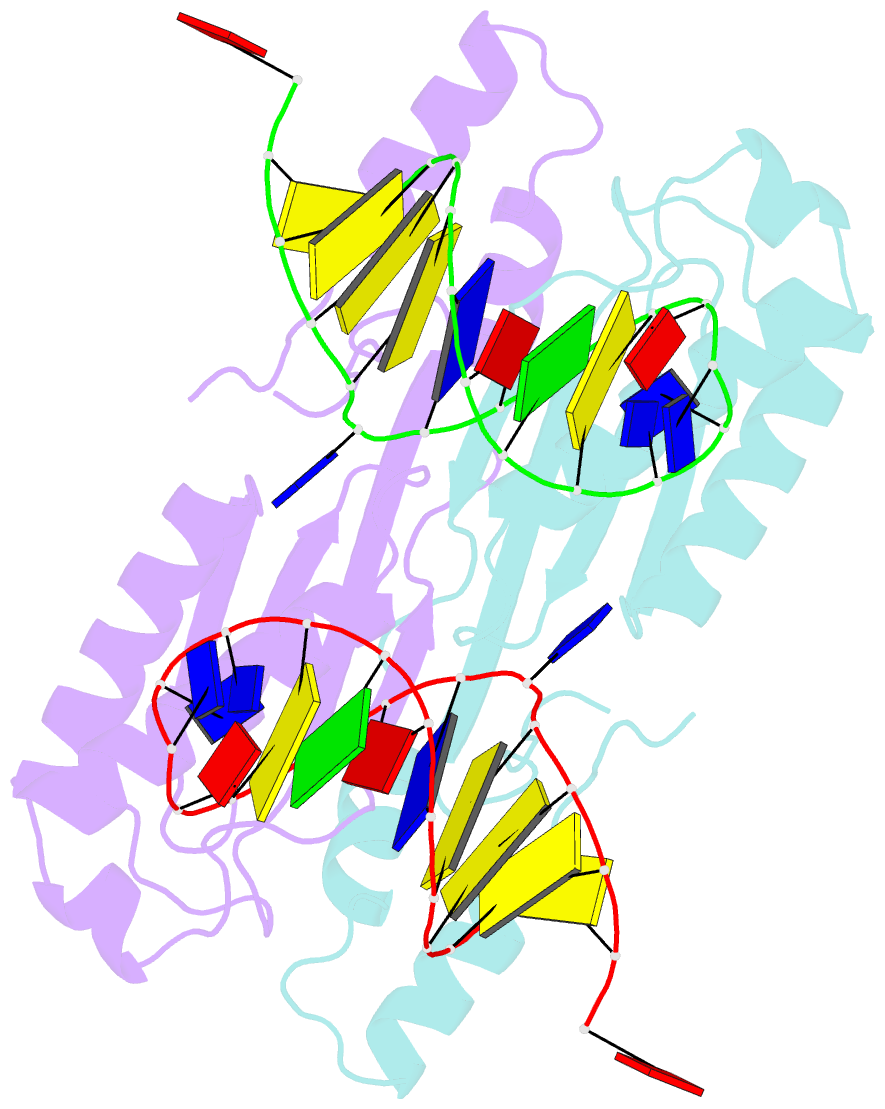
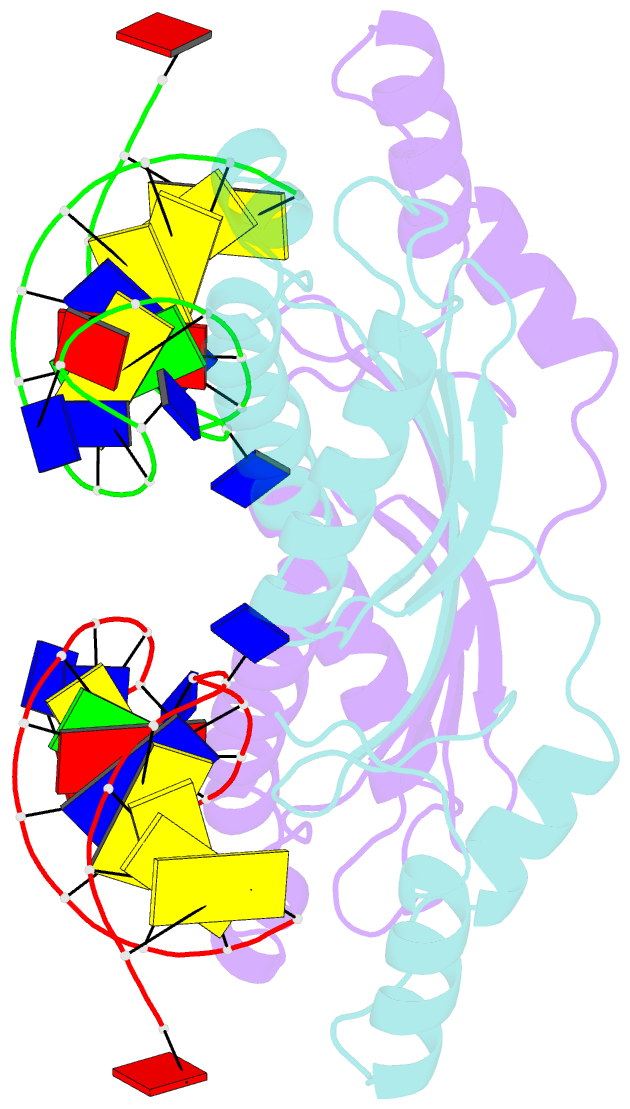
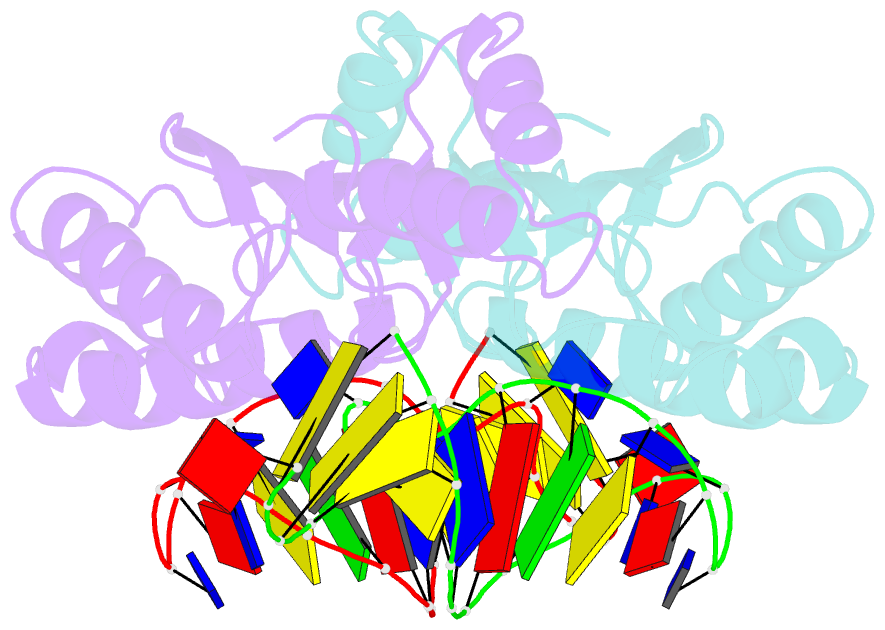
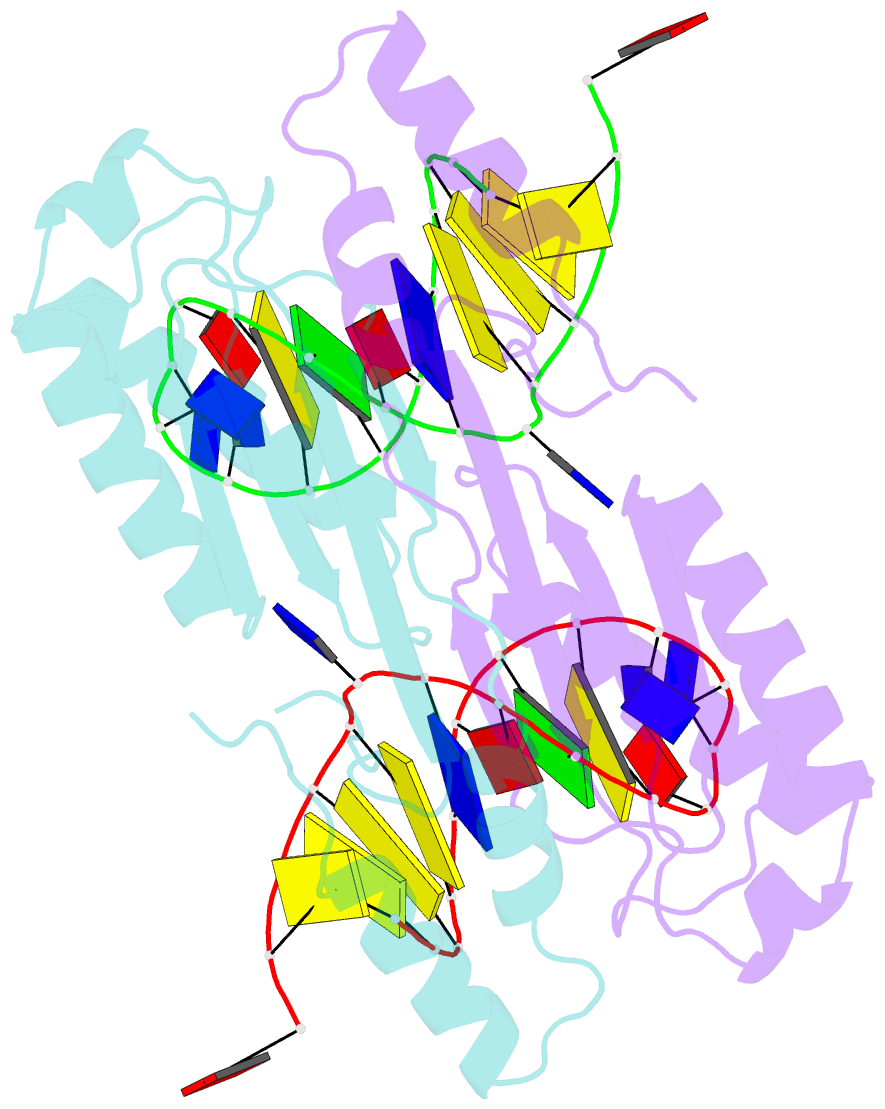
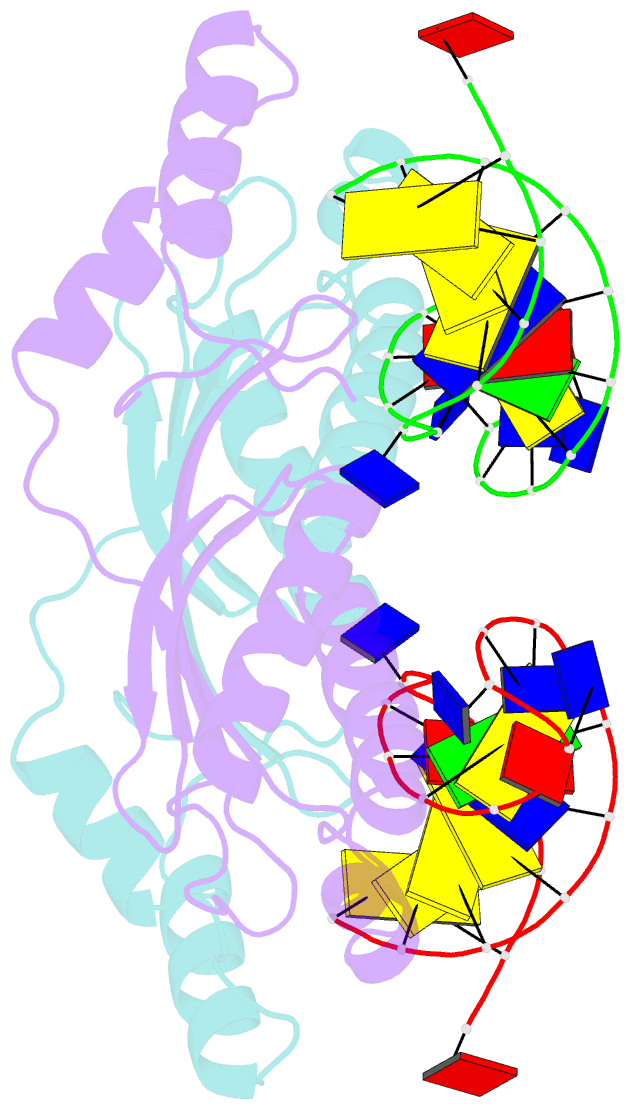
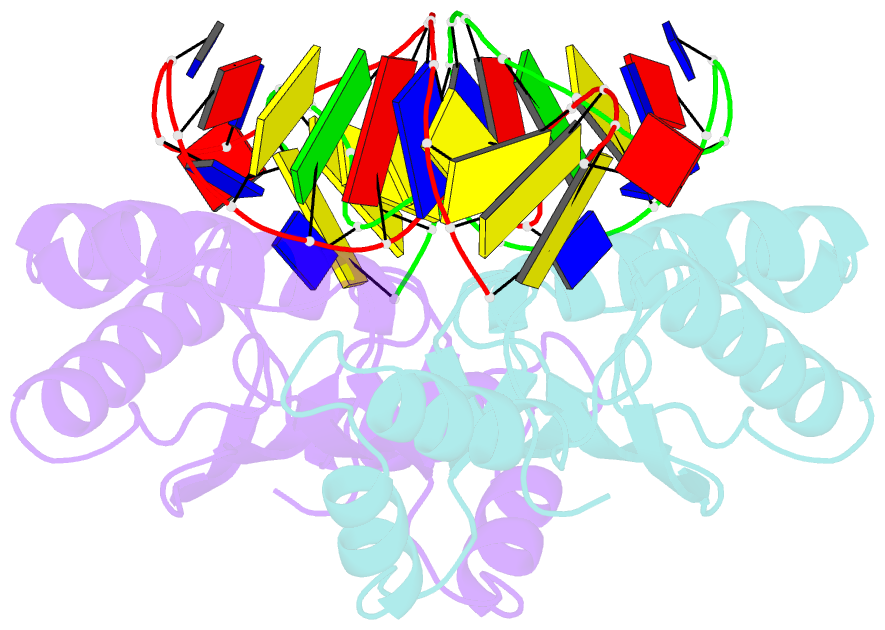
- Crystal structure of the ishp608 transposase in complex with stem-loop DNA
- Reference
- Ronning DR, Guynet C, Ton-Hoang B, Perez ZN, Ghirlando R, Chandler M, Dyda F (2005): "Active site sharing and subterminal hairpin recognition in a new class of DNA transposases." Mol.Cell, 20, 143-154. doi: 10.1016/j.molcel.2005.07.026.
- Abstract
- Many bacteria harbor simple transposable elements termed insertion sequences (IS). In Helicobacter pylori, the chimeric IS605 family elements are particularly interesting due to their proximity to genes encoding gastric epithelial invasion factors. Protein sequences of IS605 transposases do not bear the hallmarks of other well-characterized transposases. We have solved the crystal structure of full-length transposase (TnpA) of a representative member, ISHp608. Structurally, TnpA does not resemble any characterized transposase; rather, it is related to rolling circle replication (RCR) proteins. Consistent with RCR, Mg2+ and a conserved tyrosine, Tyr127, are essential for DNA nicking and the formation of a covalent intermediate between TnpA and DNA. TnpA is dimeric, contains two shared active sites, and binds two DNA stem loops representing the conserved inverted repeats near each end of ISHp608. The cocrystal structure with stem-loop DNA illustrates how this family of transposases specifically recognizes and pairs ends, necessary steps during transposition.