Summary information and primary citation
- PDB-id
- 2a8v; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- protein-RNA
- Method
- X-ray (2.4 Å)
- Summary
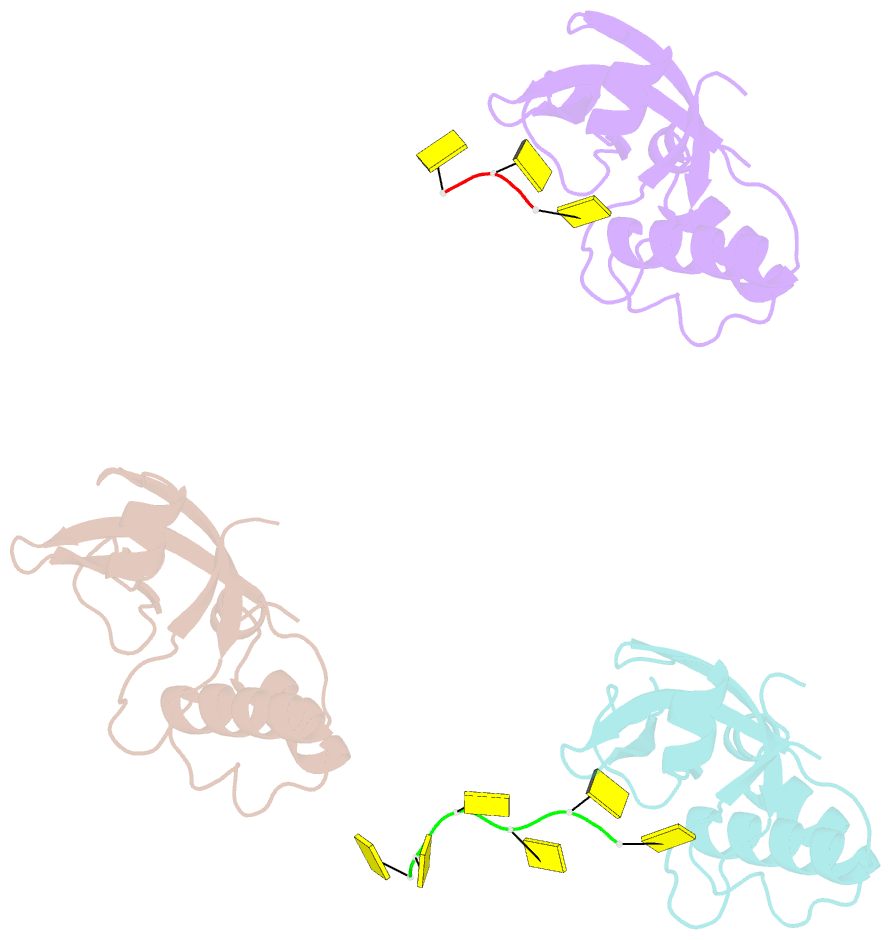
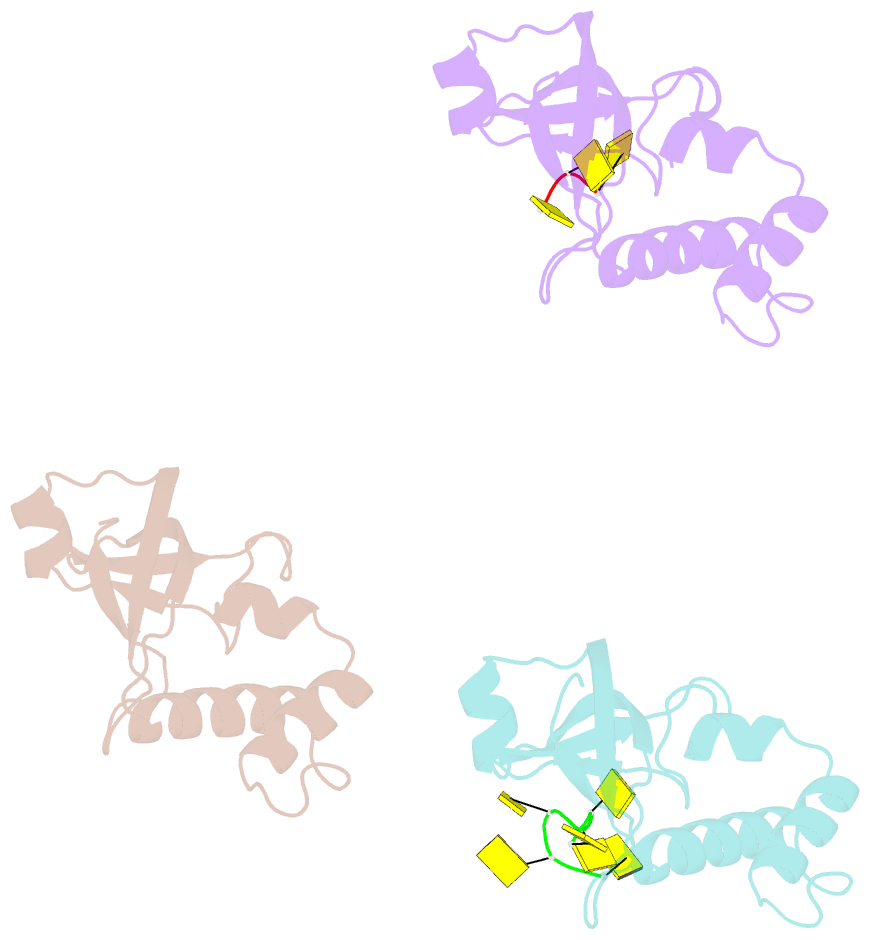
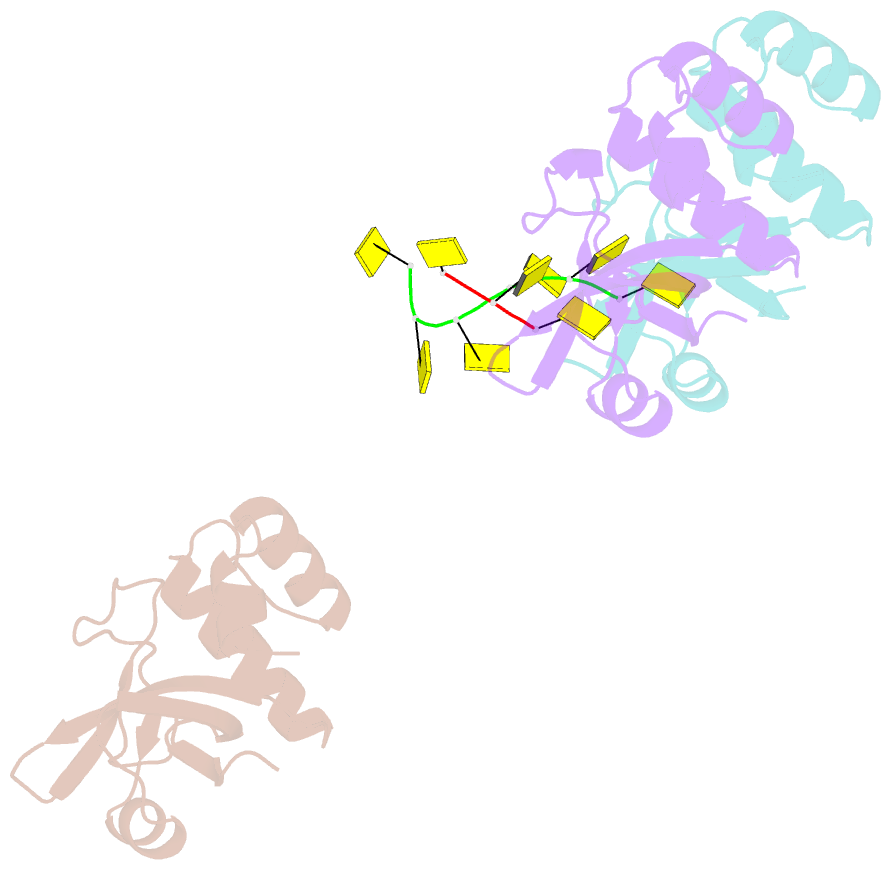
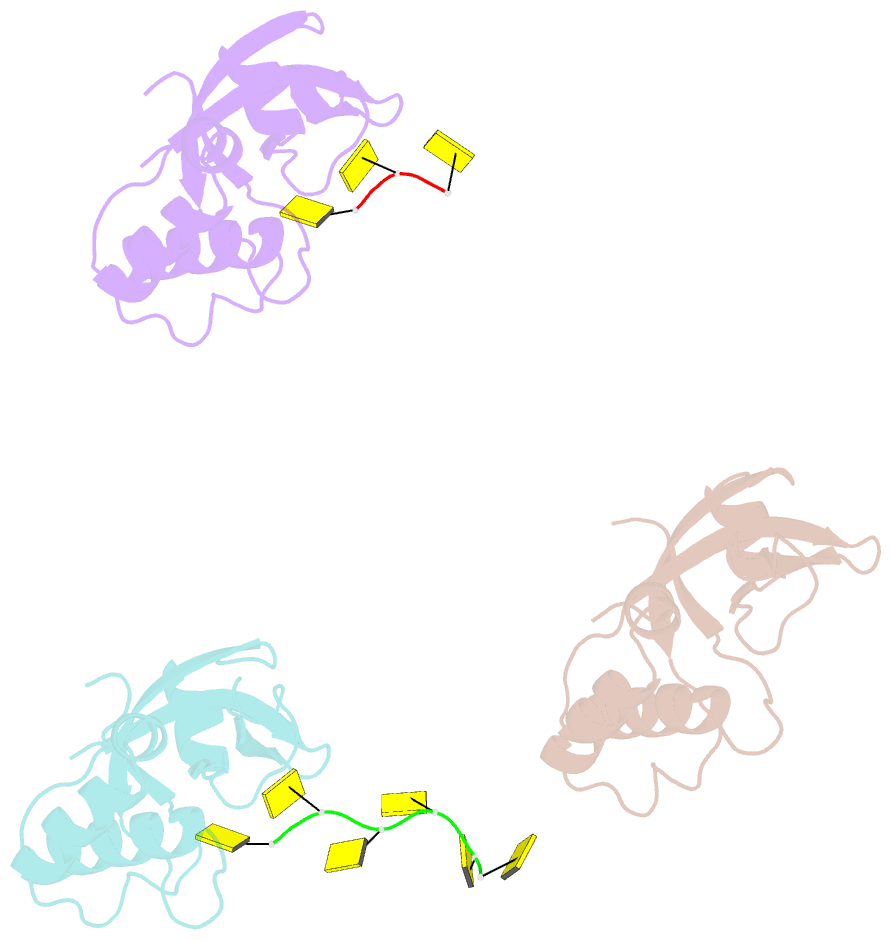
- Rho transcription termination factor-RNA complex
- Reference
- Bogden CE, Fass D, Bergman N, Nichols MD, Berger JM (1999): "The structural basis for terminator recognition by the Rho transcription termination factor." Mol.Cell, 3, 487-493. doi: 10.1016/S1097-2765(00)80476-1.
- Abstract
- The E. coli Rho protein disengages newly transcribed RNA from its DNA template, helping terminate certain transcripts. We have determined the X-ray crystal structure of the RNA-binding domain of Rho complexed to an RNA ligand. Filters that screen both ligand size and chemical functionality line the primary nucleic acid-binding site, imparting sequence specificity to a generic single-stranded nucleic acid-binding fold and explaining the preference of Rho for cytosine-rich RNA. The crystal packing reveals two Rho domain protomers bound to a single RNA with a single base spacer, suggesting that the strong RNA-binding sites of Rho may arise from pairing of RNA-binding modules. Dimerization of symmetric subunits on an asymmetric ligand is developed as a model for allosteric control in the action of the intact Rho hexamer.