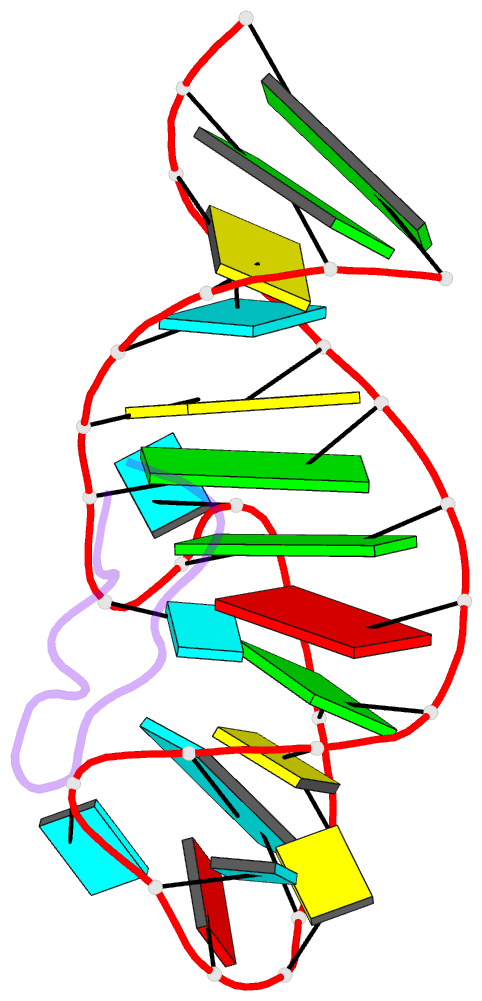
Summary information and primary citation
- PDB-id
- 2a9x; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- RNA binding protein-RNA
- Method
- NMR
- Summary
- Tar RNA recognition by a cyclic peptidomimetic of tat protein
- Reference
- Leeper TC, Athanassiou Z, Dias RL, Robinson JA, Varani G (2005): "TAR RNA recognition by a cyclic peptidomimetic of Tat protein." Biochemistry, 44, 12362-12372. doi: 10.1021/bi0510532.
- Abstract
- The search for new antiviral drugs that repress HIV viral replication by blocking transactivation of viral RNA transcription has long been advocated as an approach to novel antiviral therapy. However, research in this area has so far failed to yield attractive lead compounds because of the insufficient development of RNA-based medicinal chemistry. One difficulty in efforts to inhibit protein-RNA interactions using small druglike molecules is the large surface areas typically found at these interfaces. To overcome this problem, we sought to identify constrained peptidomimetic inhibitors that would provide potential new drug leads. We previously reported the discovery of a cyclic peptide mimic of the RNA-binding domain of BIV Tat protein based on a designed beta-hairpin scaffold. We demonstrated that the cyclic peptide bound BIV TAR RNA with an affinity comparable to that of the RNA-binding domain of the Tat protein and inhibited protein binding to the RNA. In this study, we report the structure of the complex of the cyclic peptide bound to BIV TAR RNA determined using heteronuclear NMR methods. The structure reveals a beta-hairpin conformation in the bound peptide, which adopts an unexpected orientation in the major groove of the RNA opposite those observed for peptides derived from the Tat protein. This structure suggests many ways in which to optimize the compound and enhance its activity and pharmacological potential and represents a further step in the rational design of a new class of HIV-1 virus replication inhibitors based on peptidomimetic chemistry.