Summary information and primary citation
- PDB-id
- 2ad9; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- RNA binding protein-RNA
- Method
- NMR
- Summary
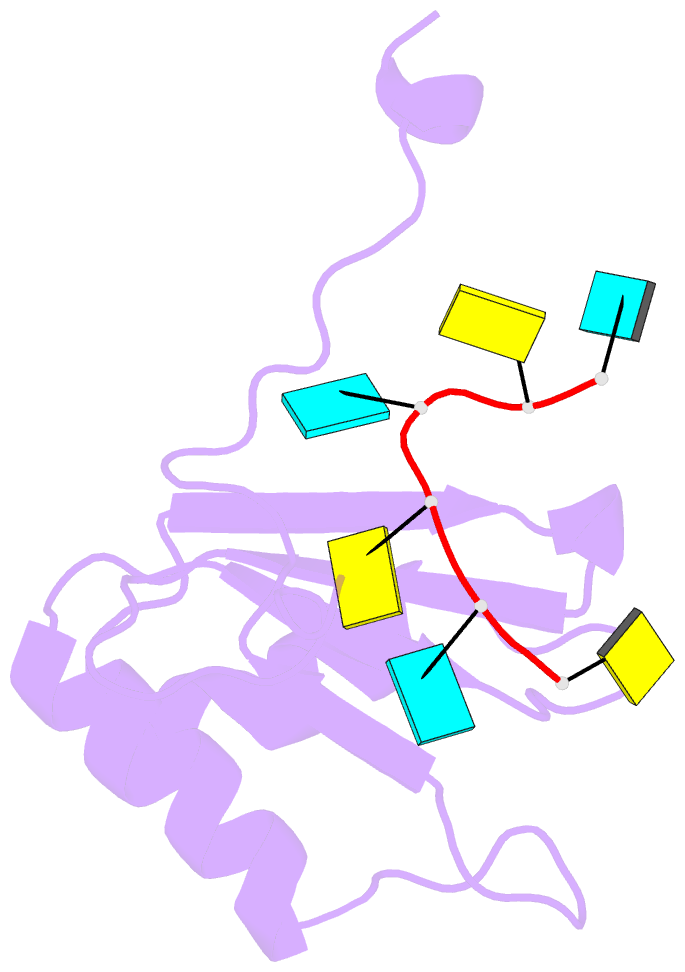
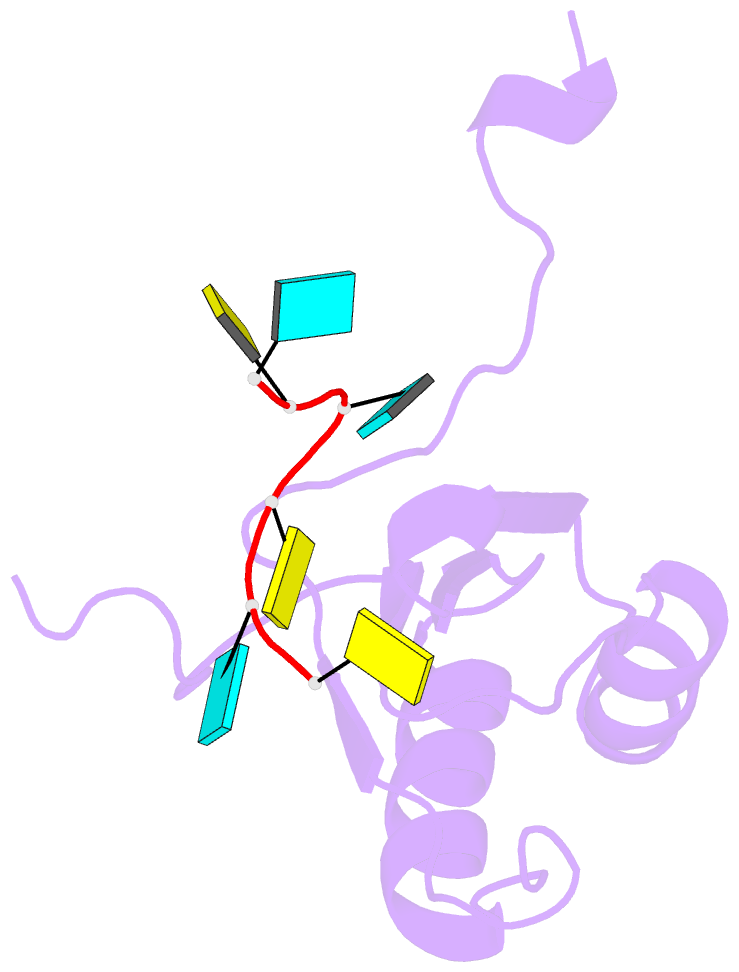
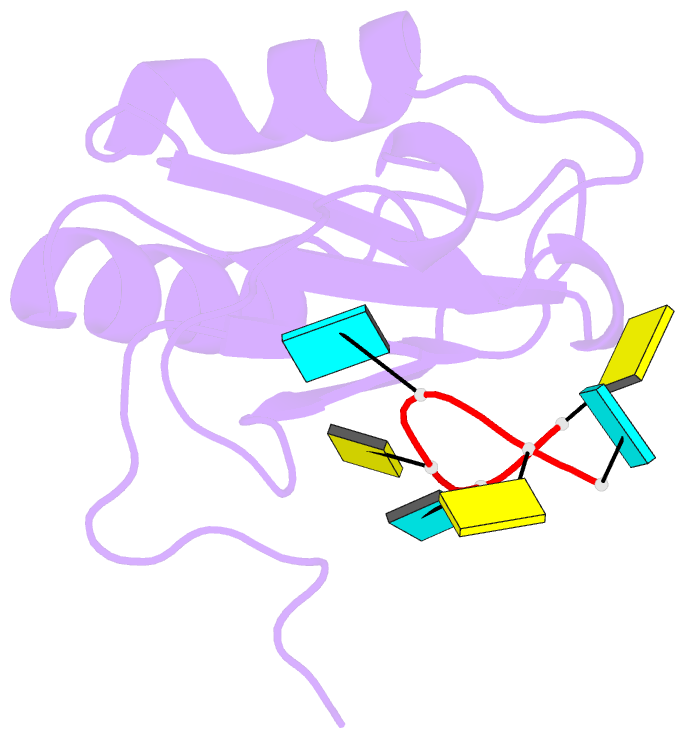
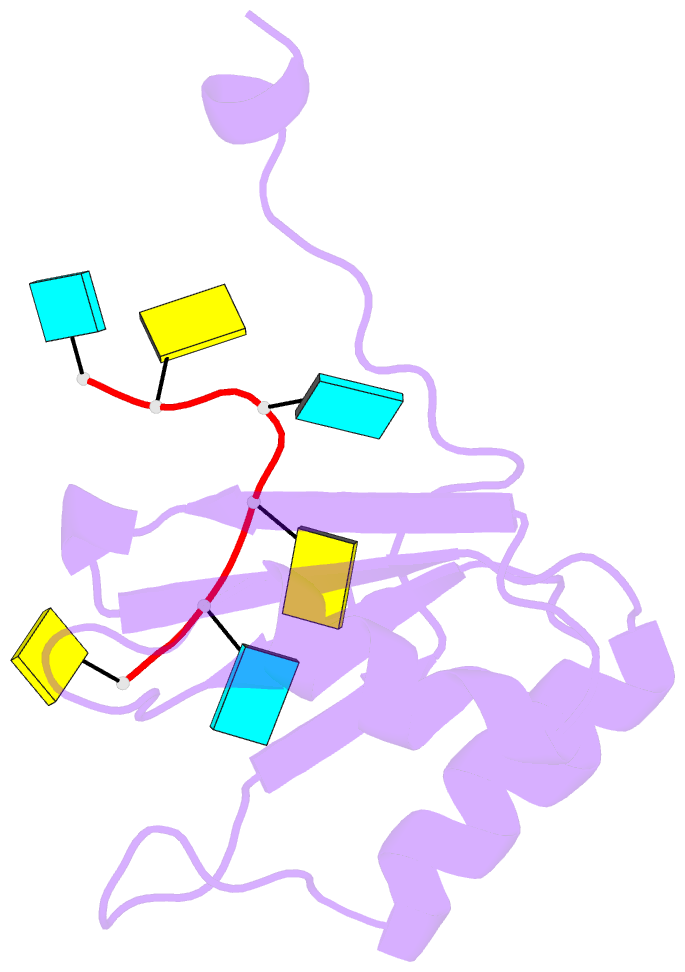
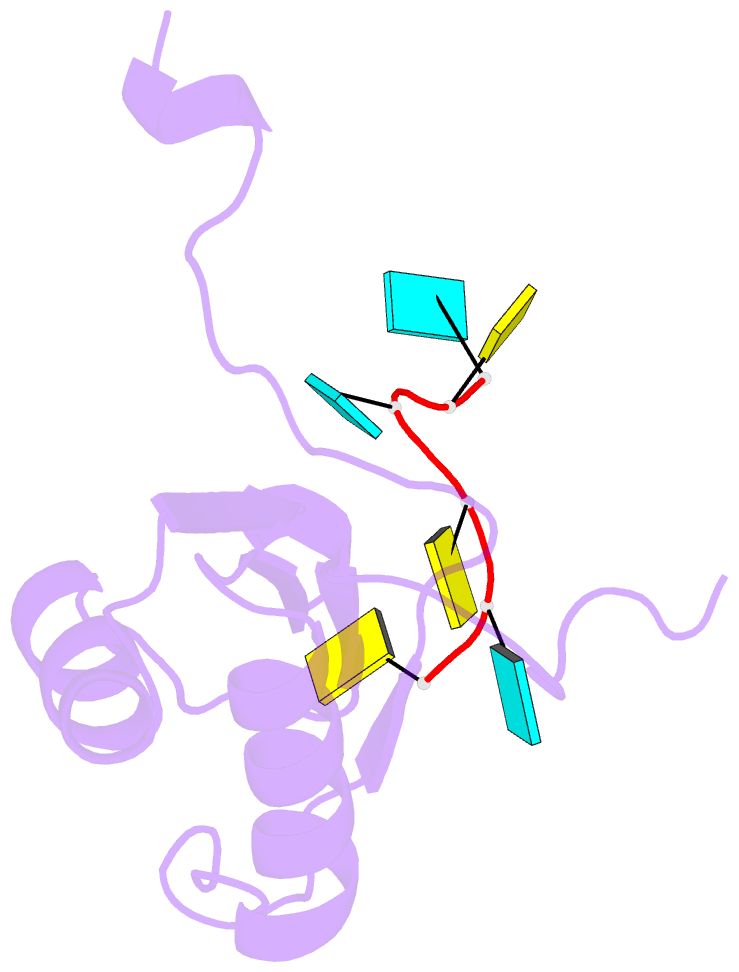
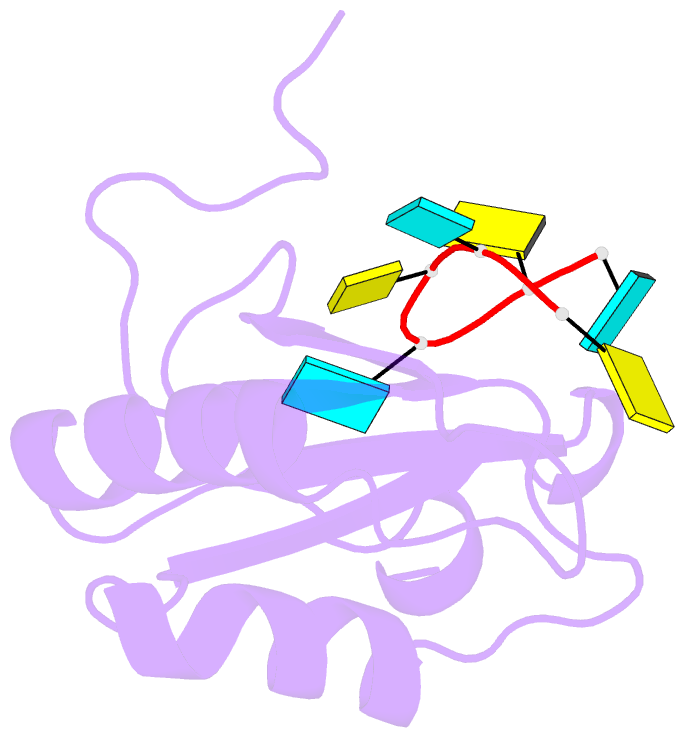
- Solution structure of polypyrimidine tract binding protein rbd1 complexed with cucucu RNA
- Reference
- Oberstrass FC, Auweter SD, Erat M, Hargous Y, Henning A, Wenter P, Reymond L, Amir-Ahmady B, Pitsch S, Black DL, Allain FHT (2005): "Structure of PTB bound to RNA: specific binding and implications for splicing regulation." Science, 309, 2054-2057. doi: 10.1126/science.1114066.
- Abstract
- The polypyrimidine tract binding protein (PTB) is a 58-kilodalton RNA binding protein involved in multiple aspects of messenger RNA metabolism, including the repression of alternative exons. We have determined the solution structures of the four RNA binding domains (RBDs) of PTB, each bound to a CUCUCU oligonucleotide. Each RBD binds RNA with a different binding specificity. RBD3 and RBD4 interact, resulting in an antiparallel orientation of their bound RNAs. Thus, PTB will induce RNA looping when bound to two separated pyrimidine tracts within the same RNA. This leads to structural models for how PTB functions as an alternative-splicing repressor.