Summary information and primary citation
- PDB-id
- 2alz; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transferase-DNA
- Method
- X-ray (2.5 Å)
- Summary
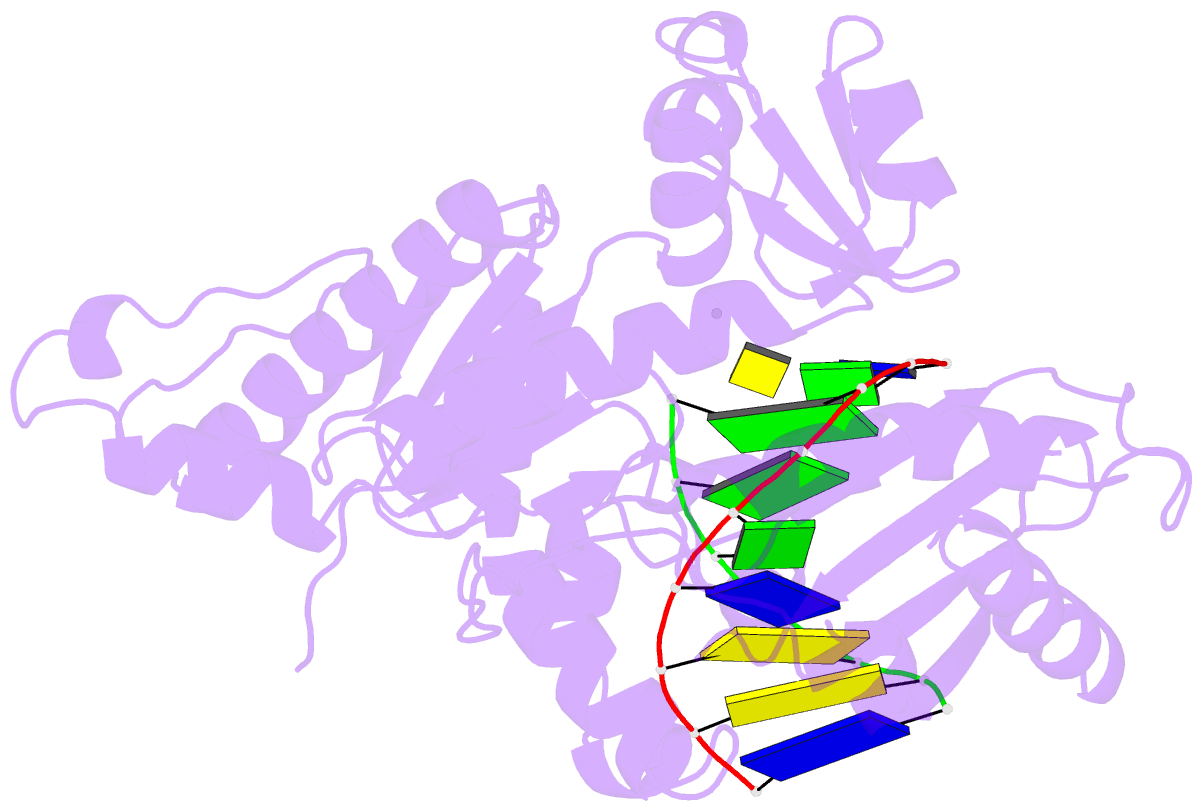
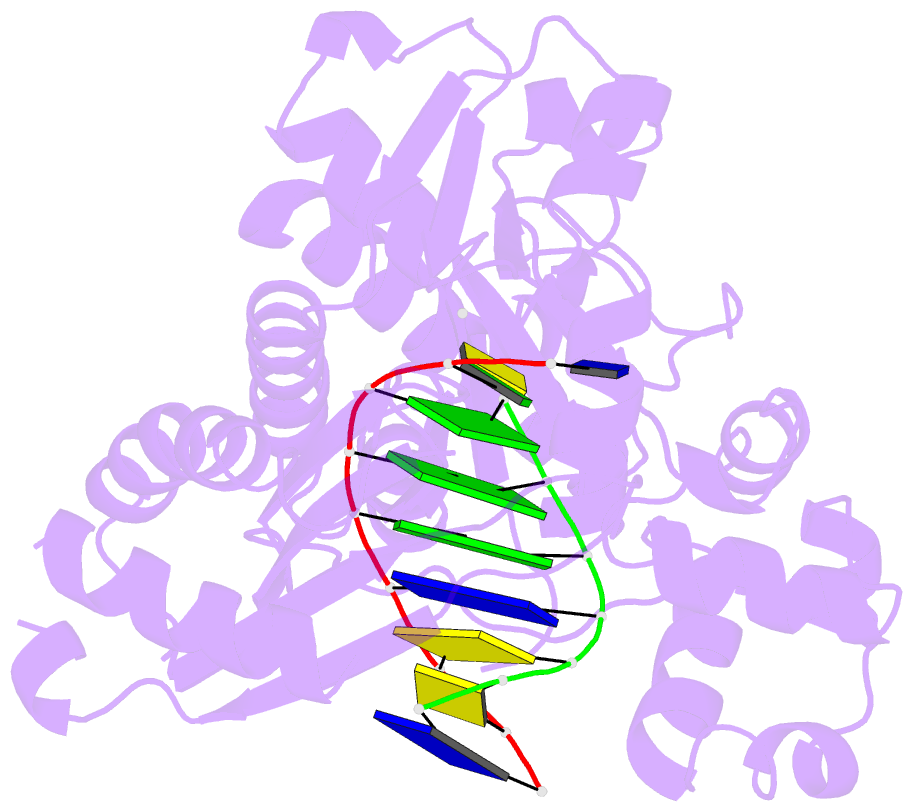
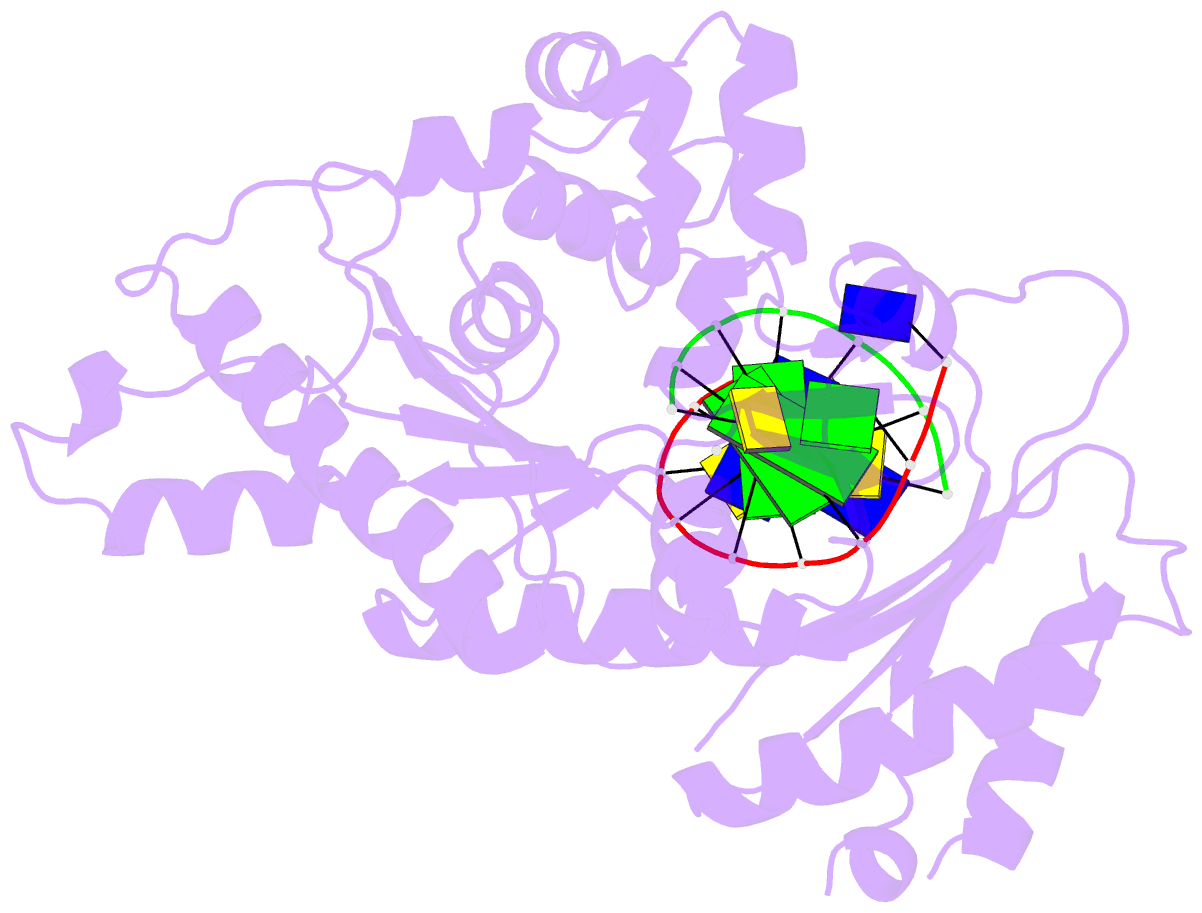
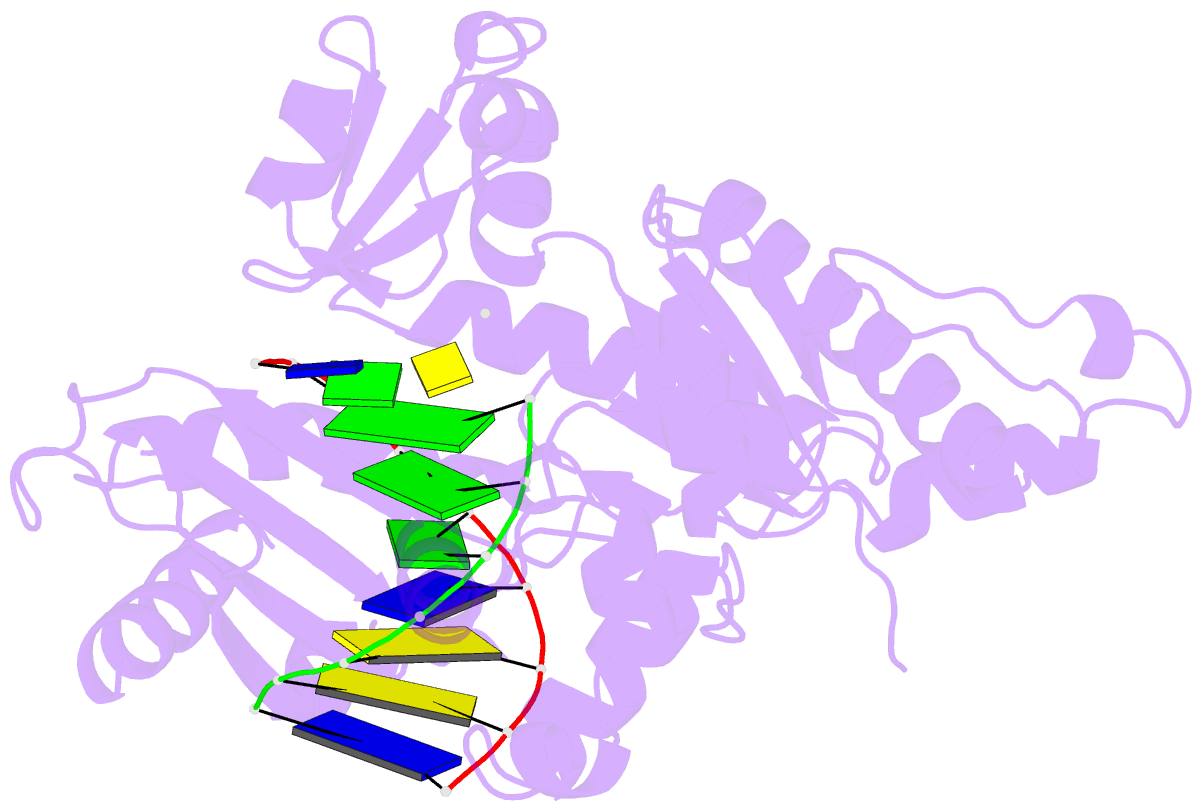
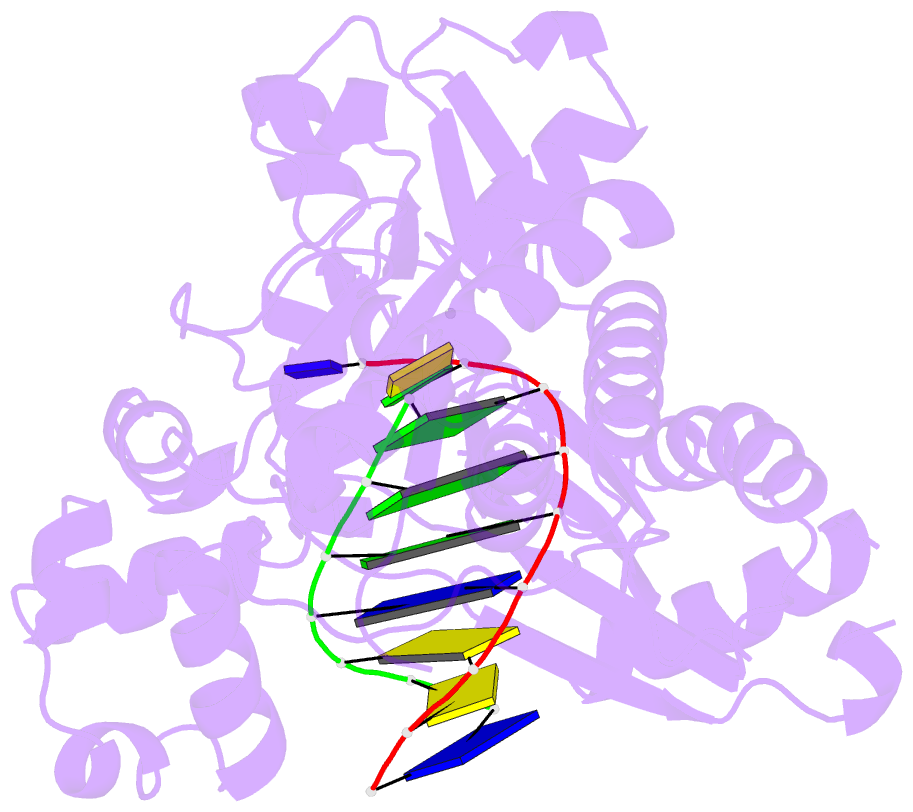
- Ternary complex of hpoli with DNA and dctp
- Reference
- Nair DT, Johnson RE, Prakash L, Prakash S, Aggarwal AK (2005): "Human DNA Polymerase iota Incorporates dCTP Opposite Template G via a G.C+ Hoogsteen Base Pair." Structure, 13, 1569-1577. doi: 10.1016/j.str.2005.08.010.
- Abstract
- Human DNA polymerase iota (hPoliota), a member of the Y family of DNA polymerases, differs in remarkable ways from other DNA polymerases, incorporating correct nucleotides opposite template purines with a much higher efficiency and fidelity than opposite template pyrimidines. We present here the crystal structure of hPoliota bound to template G and incoming dCTP, which reveals a G.C + Hoogsteen base pair in a DNA polymerase active site. We show that the hPoliota active site has evolved to favor Hoogsteen base pairing, wherein the template sugar is fixed in a cavity that reduces the C1'-C1' distance across the nascent base pair from approximately 10.5 A in other DNA polymerases to 8.6 A in hPoliota. The rotation of G from anti to syn is then largely in response to this curtailed C1'-C1' distance. A G.C+ Hoogsteen base pair suggests a specific mechanism for hPoliota's ability to bypass N(2)-adducted guanines that obstruct replication.