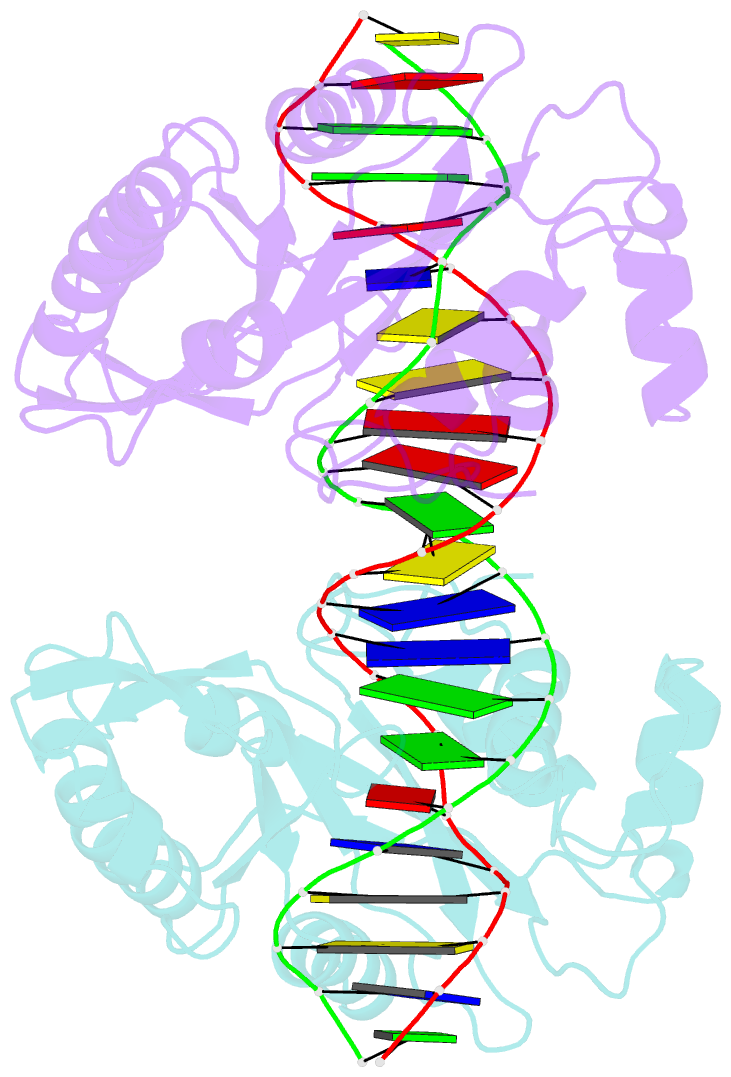
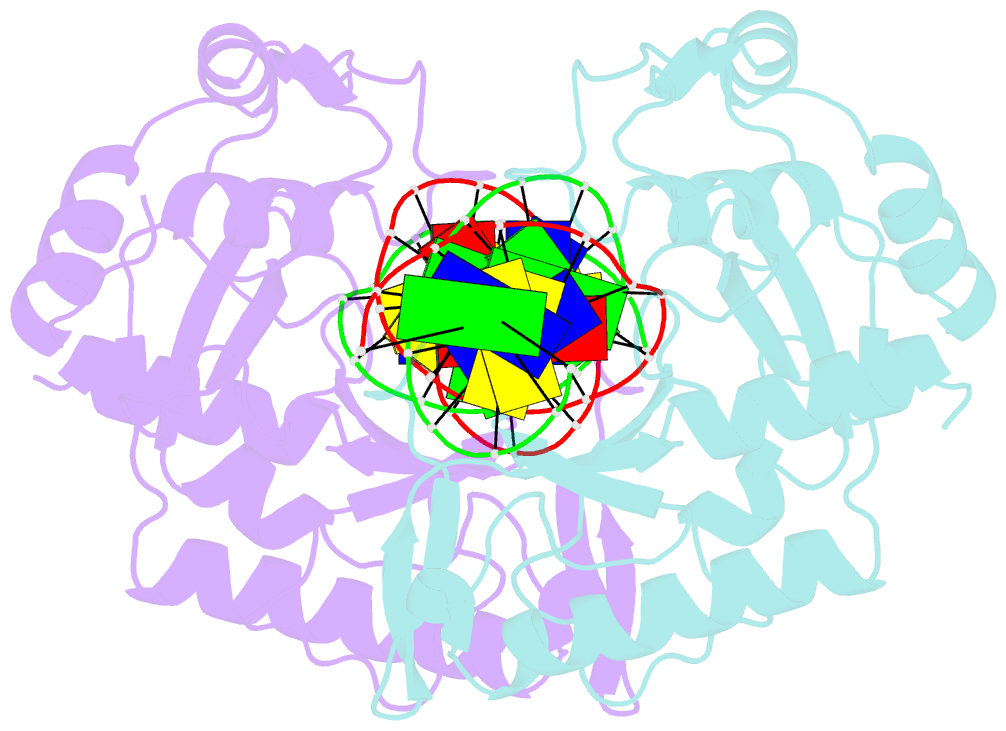
Summary information and primary citation
- PDB-id
- 2aor; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- hydrolase-DNA
- Method
- X-ray (2.0 Å)
- Summary
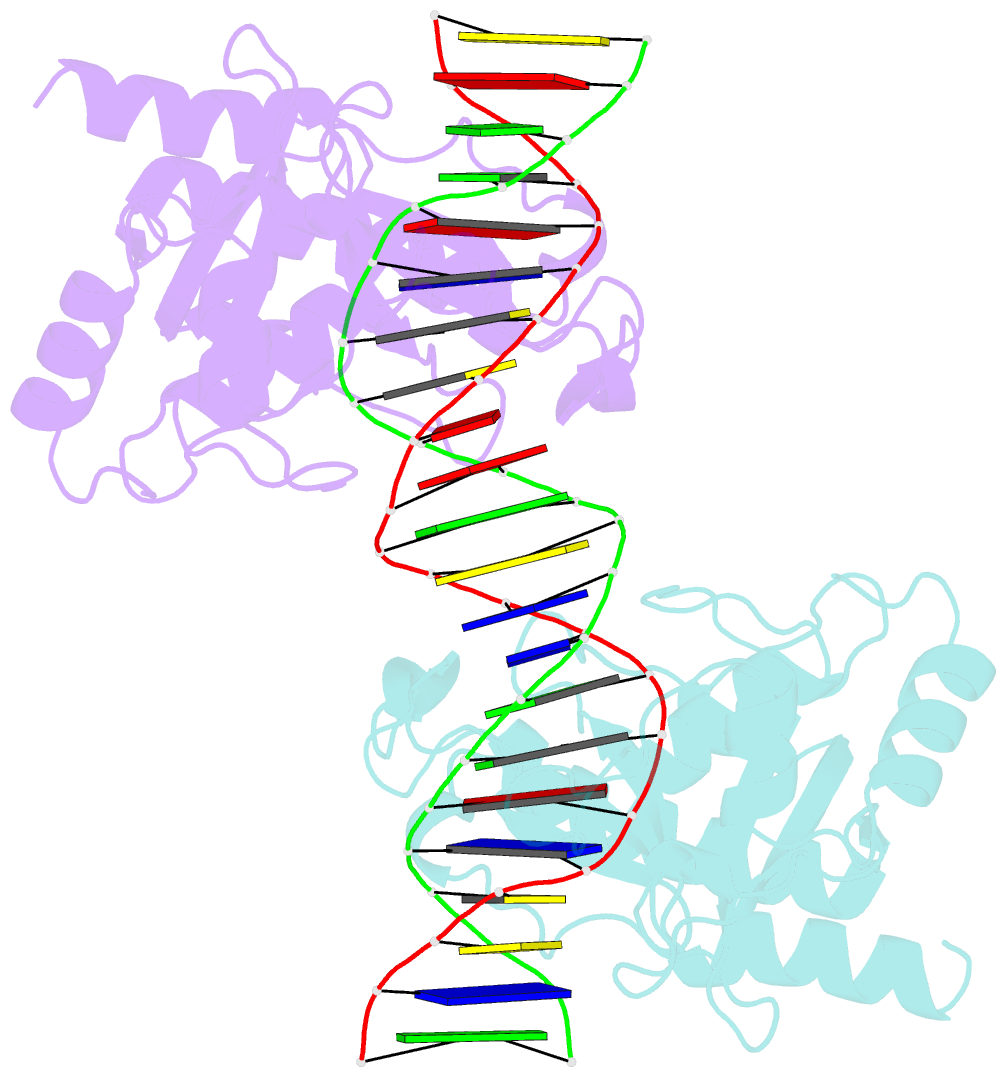
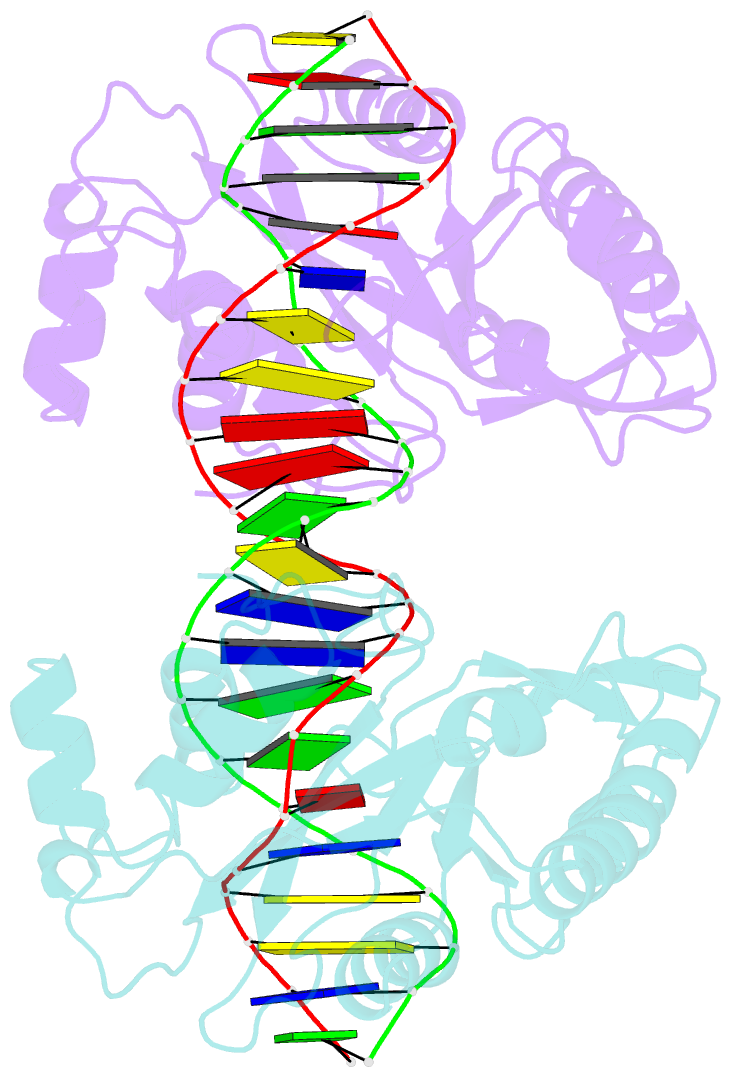
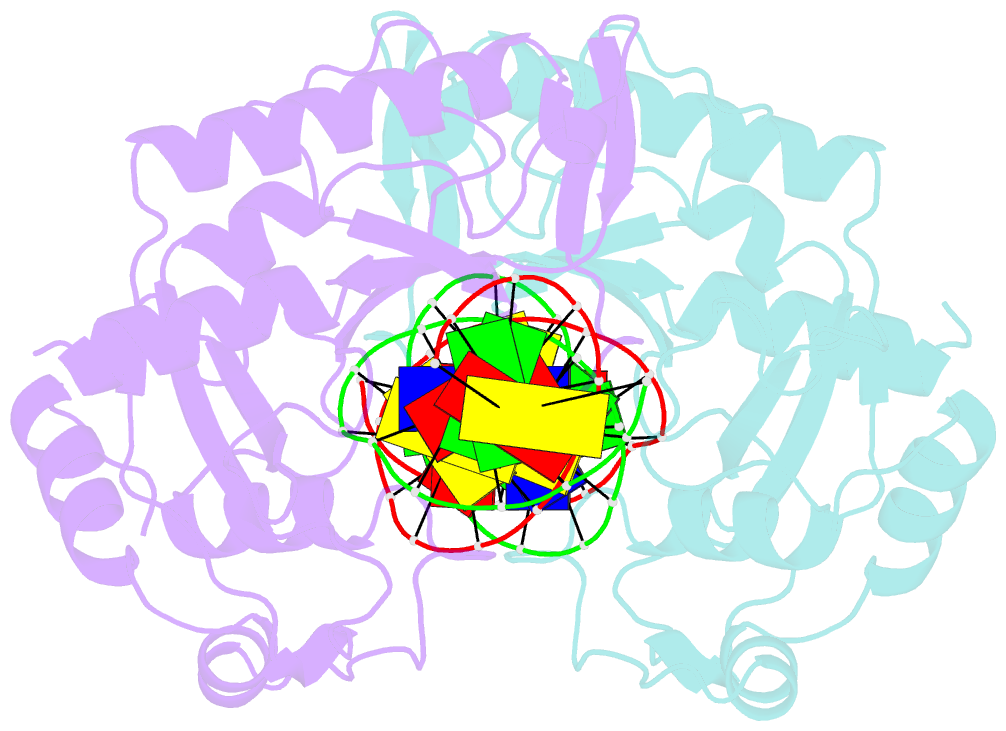
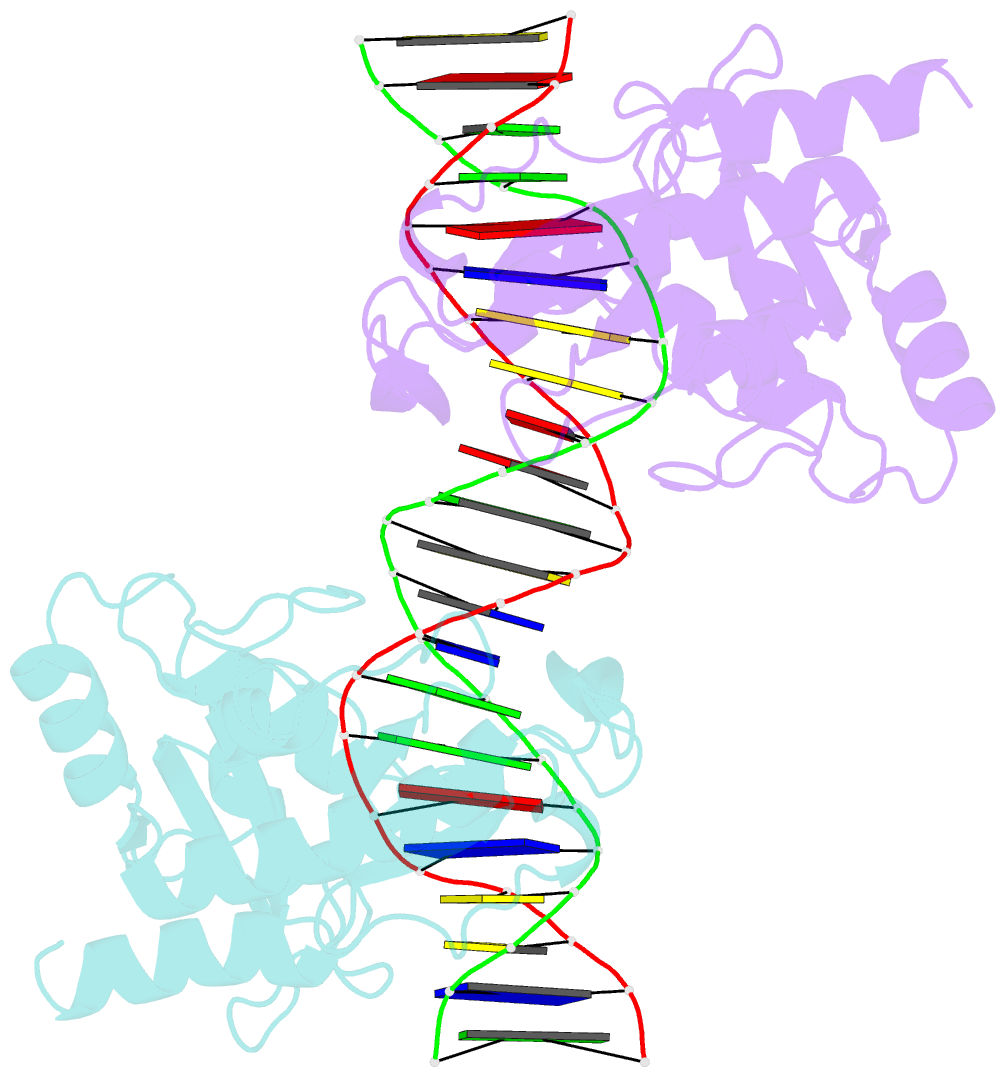
- Crystal structure of muth-hemimethylated DNA complex
- Reference
- Lee JY, Chang J, Joseph N, Ghirlando R, Rao DN, Yang W (2005): "MutH complexed with hemi- and unmethylated DNAs: coupling base recognition and DNA cleavage." Mol.Cell, 20, 155-166. doi: 10.1016/j.molcel.2005.08.019.
- Abstract
- MutH initiates mismatch repair by nicking the transiently unmethylated daughter strand 5' to a GATC sequence. Here, we report crystal structures of MutH complexed with hemimethylated and unmethylated GATC substrates. Both structures contain two Ca2+ ions jointly coordinated by a conserved aspartate and the scissile phosphate, as observed in the restriction endonucleases BamHI and BglI. In the hemimethylated complexes, the active site is more compact and DNA cleavage is more efficient. The Lys residue in the conserved DEK motif coordinates the nucleophilic water in conjunction with the phosphate 3' to the scissile bond; the same Lys is also hydrogen bonded with a carbonyl oxygen in the DNA binding module. We propose that this Lys, which is conserved in many restriction endonucleases and is replaced by Glu or Gln in BamHI and BglII, is a sensor for DNA binding and the linchpin that couples base recognition and DNA cleavage.