Summary information and primary citation
- PDB-id
- 2aq4; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transferase
- Method
- X-ray (2.32 Å)
- Summary
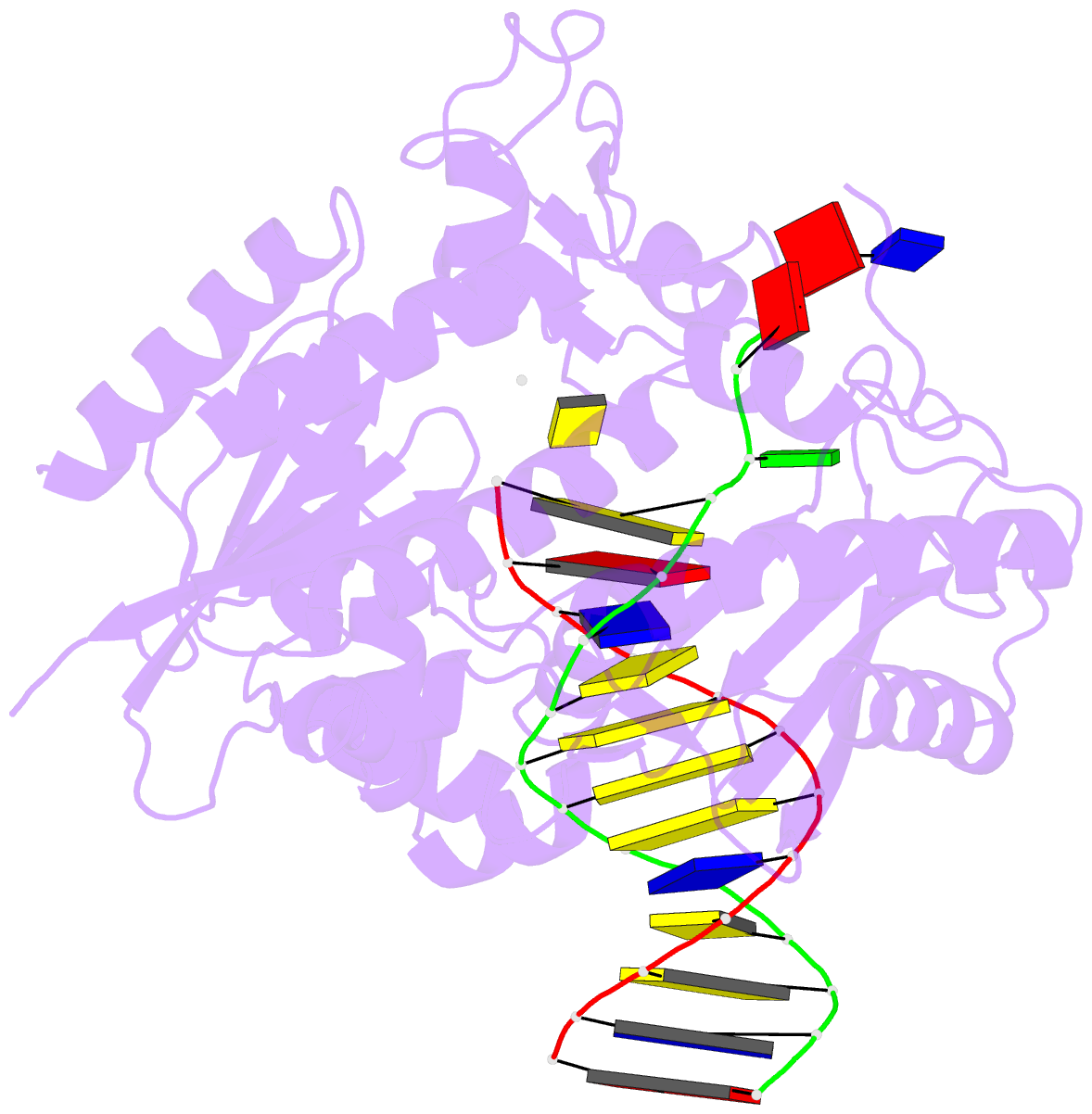
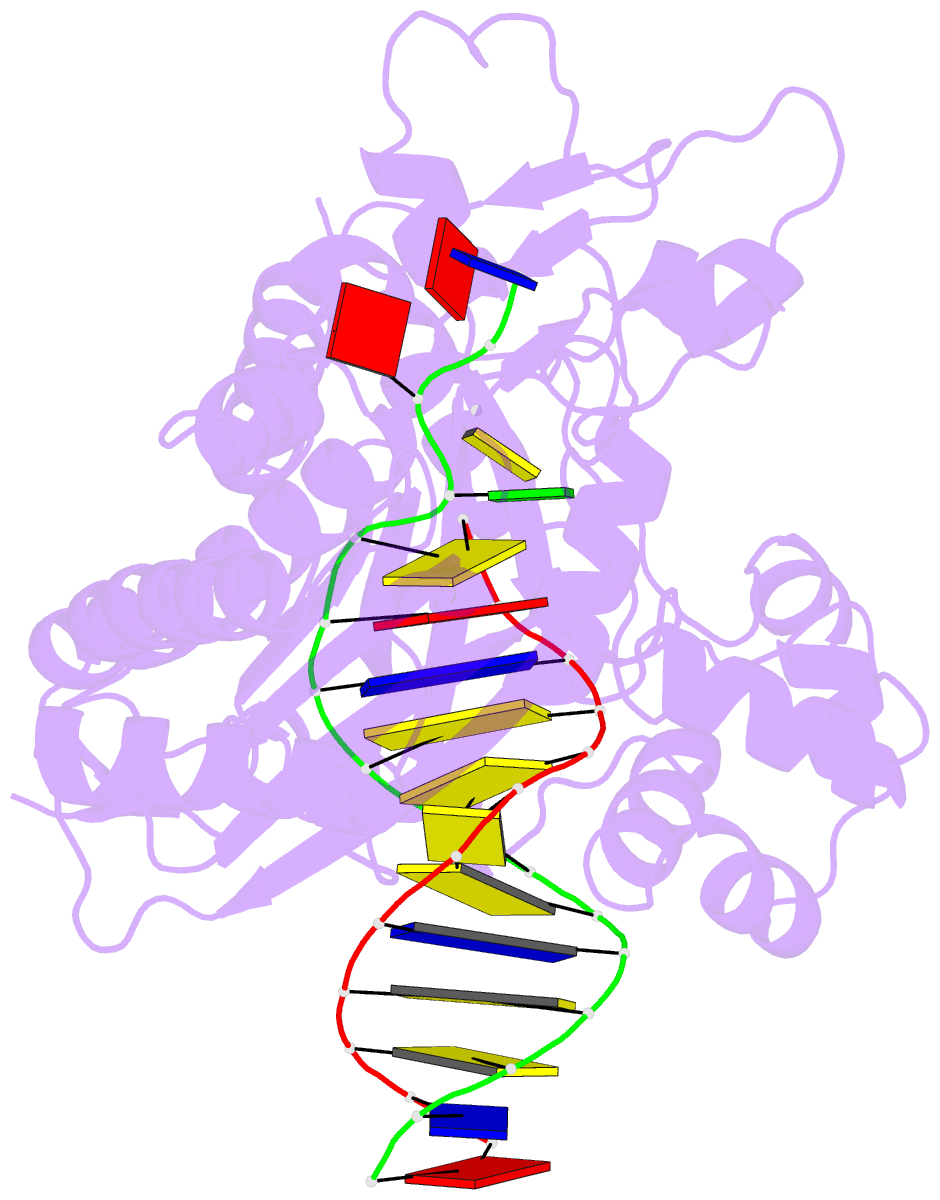
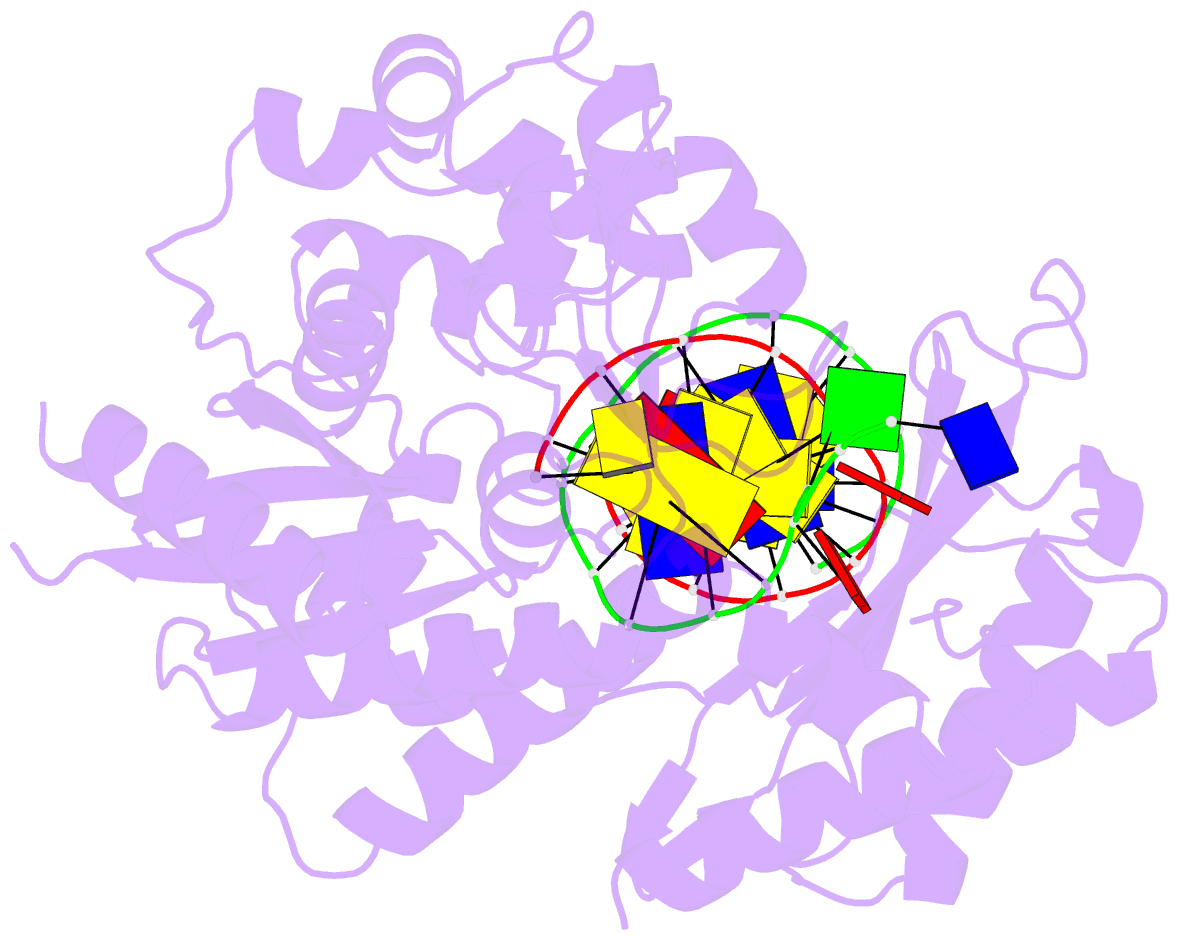
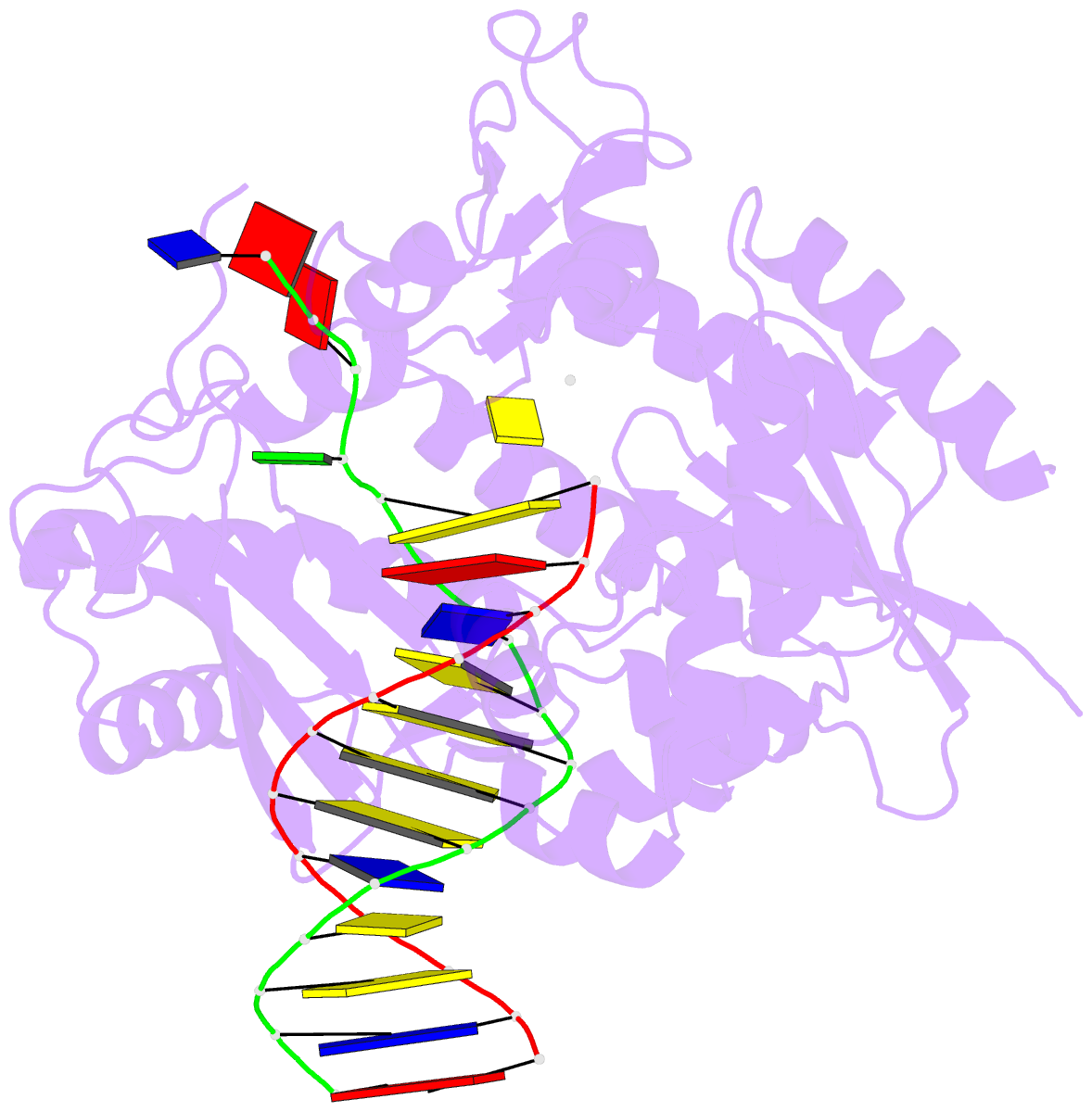
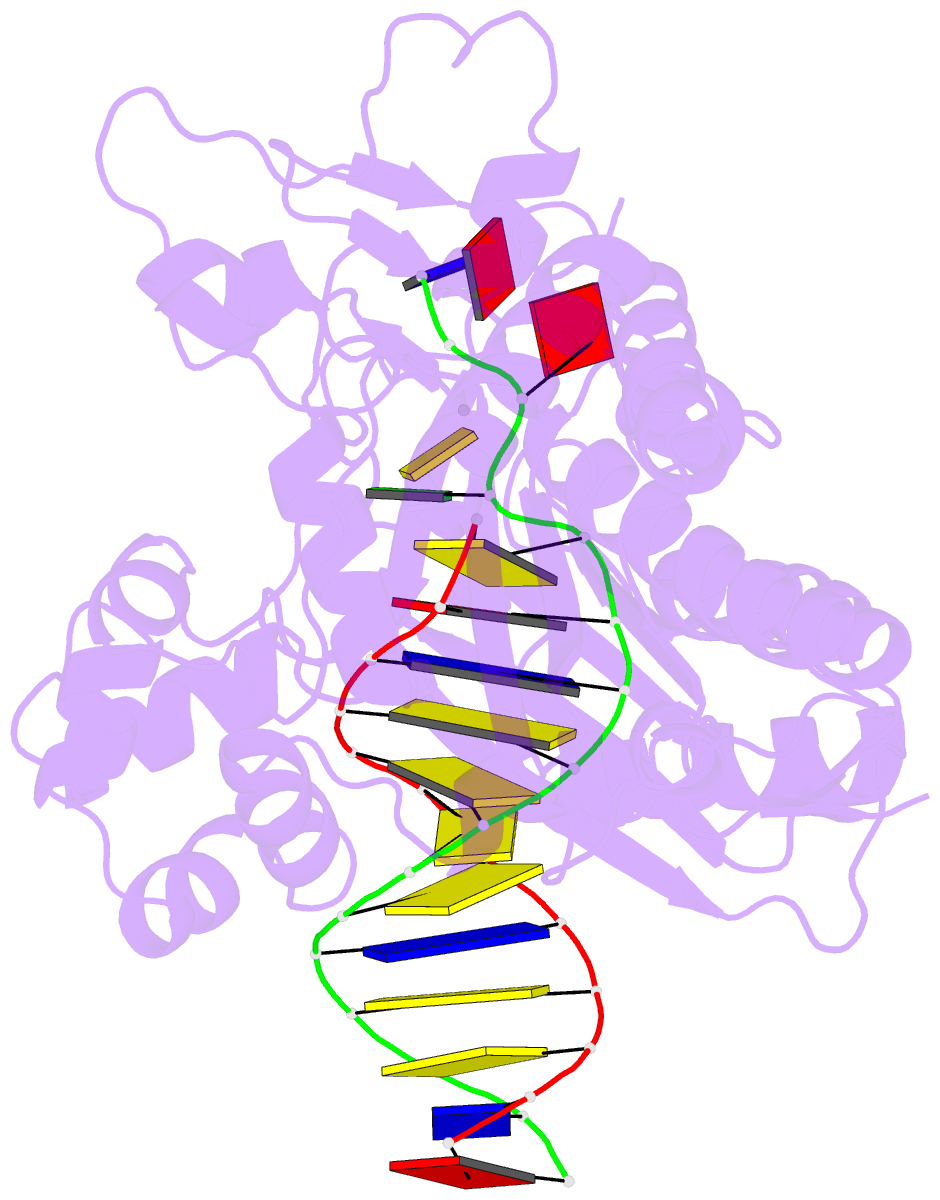
- Ternary complex of the catalytic core of rev1 with DNA and dctp.
- Reference
- Nair DT, Johnson RE, Prakash L, Prakash S, Aggarwal AK (2005): "Rev1 employs a novel mechanism of DNA synthesis using a protein template." Science, 309, 2219-2222. doi: 10.1126/science.1116336.
- Abstract
- The Rev1 DNA polymerase is highly specialized for the incorporation of C opposite template G. We present here the crystal structure of yeast Rev1 bound to template G and incoming 2'-deoxycytidine 5'-triphosphate (dCTP), which reveals that the polymerase itself dictates the identity of the incoming nucleotide, as well as the identity of the templating base. Template G and incoming dCTP do not pair with each other. Instead, the template G is evicted from the DNA helix, and it makes optimal hydrogen bonds with a segment of Rev1. Also, unlike other DNA polymerases, incoming dCTP pairs with an arginine rather than the templating base, which ensures the incorporation of dCTP over other incoming nucleotides. This mechanism provides an elegant means for promoting proficient and error-free synthesis through N2-adducted guanines that obstruct replication.