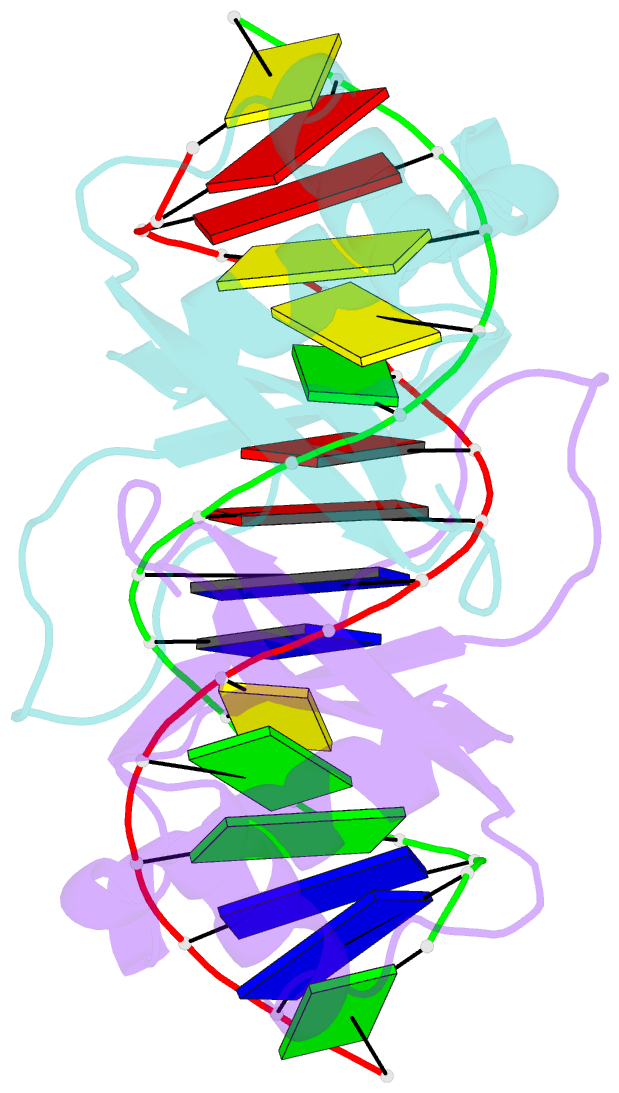
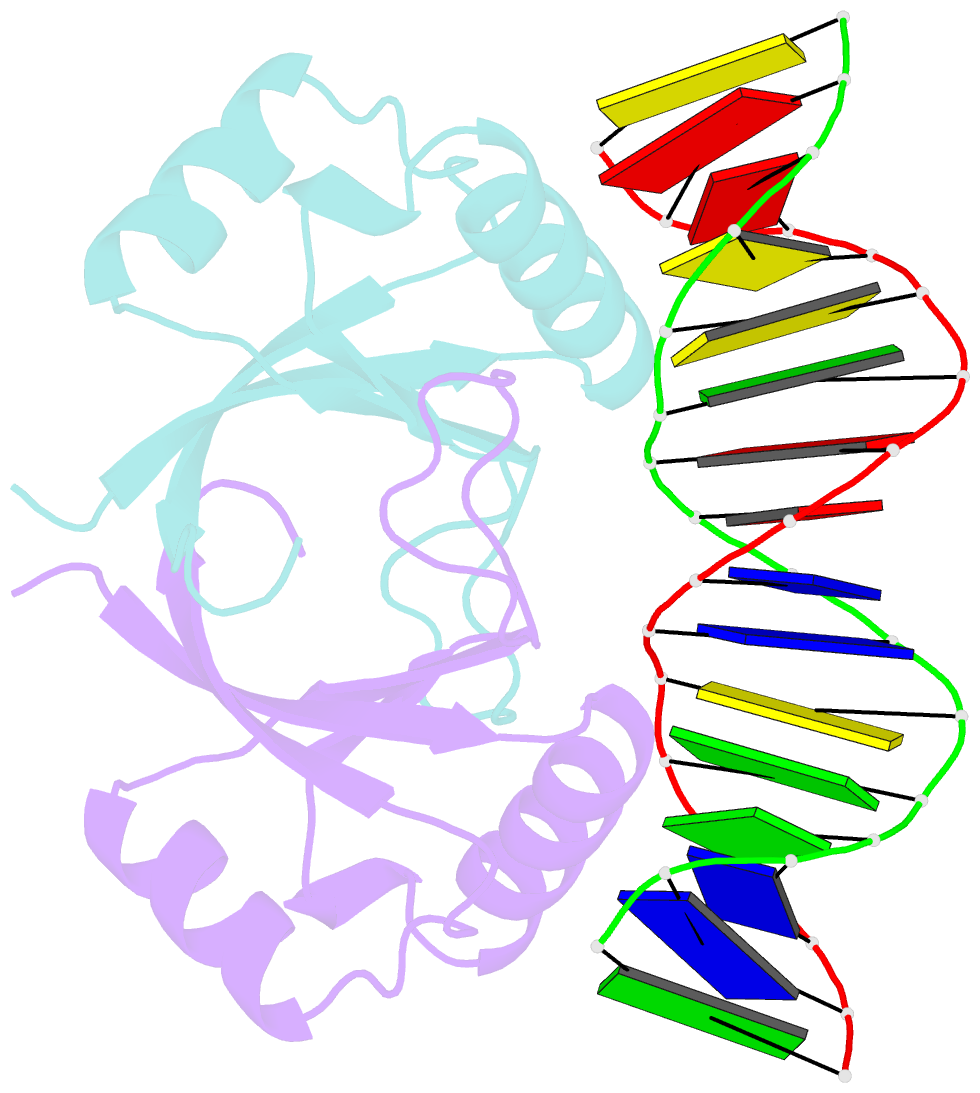
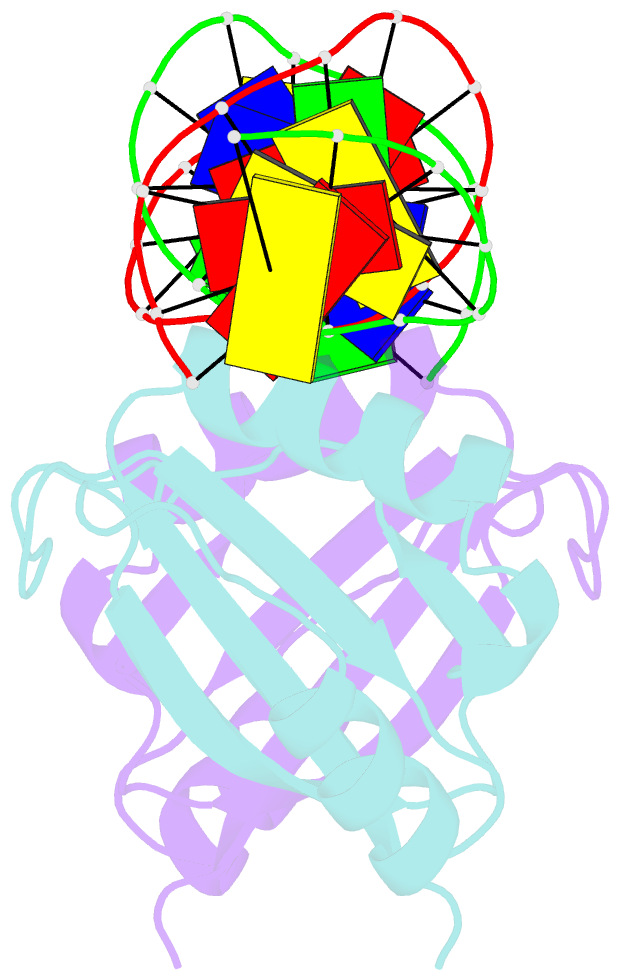
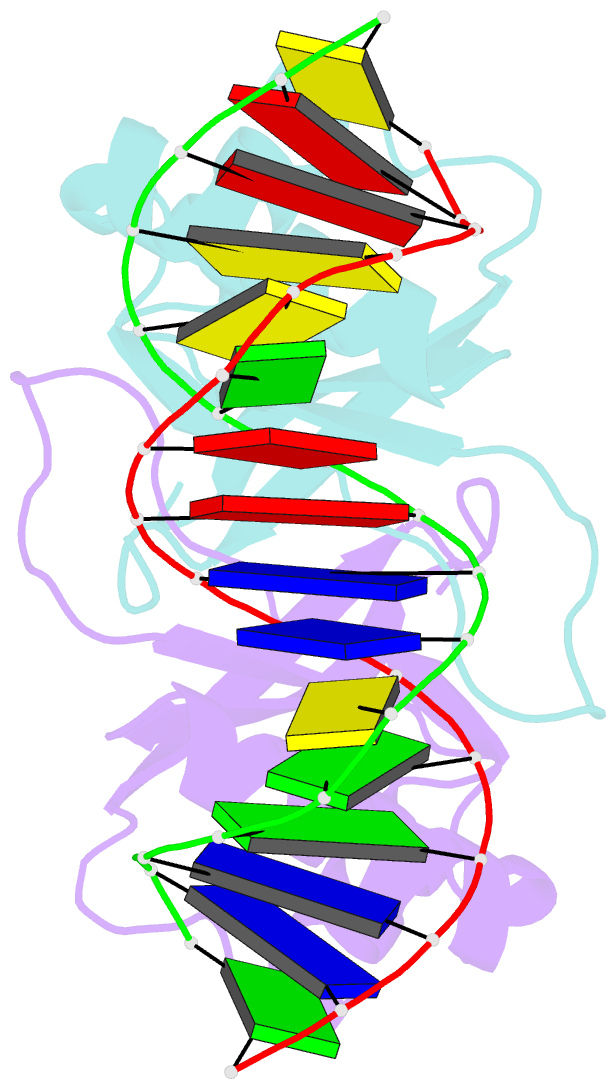
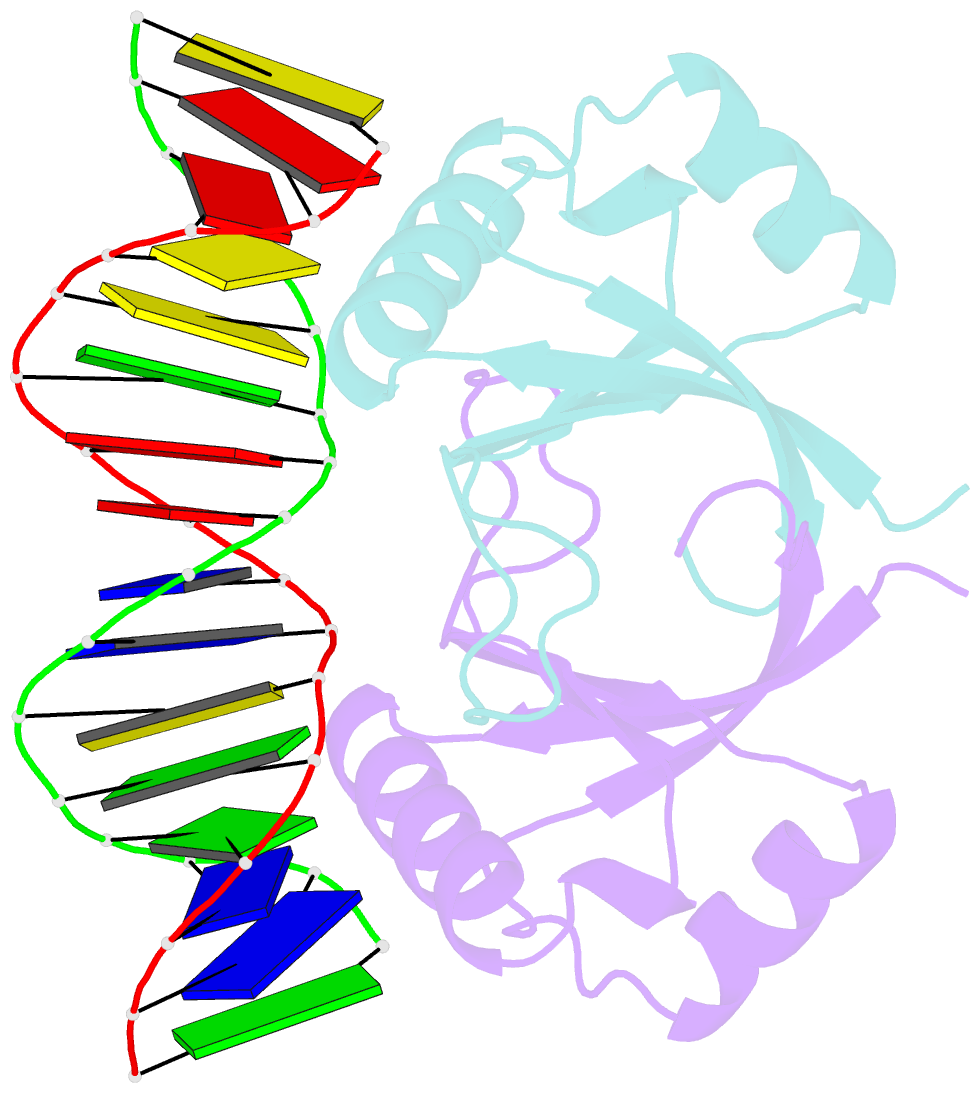
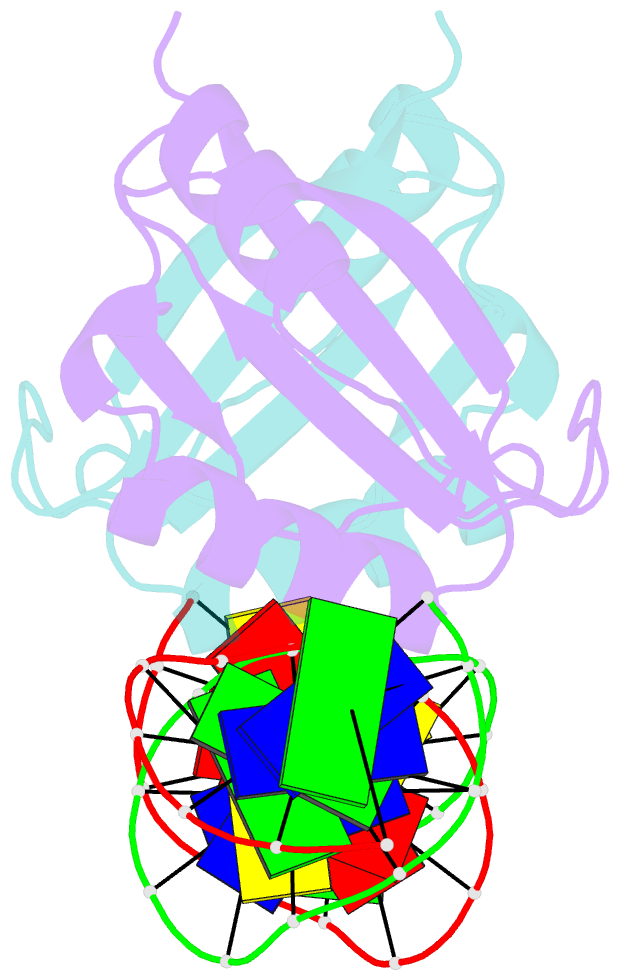
Summary information and primary citation
- PDB-id
- 2ayb; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transcription-DNA
- Method
- X-ray (3.2 Å)
- Summary
- Crystal structure of hpv6a e2 DNA binding domain bound to a 16 base pair DNA target
- Reference
- Hooley E, Fairweather V, Clarke AR, Gaston K, Brady RL (2006): "The recognition of local DNA conformation by the human papillomavirus type 6 E2 protein." Nucleic Acids Res., 34, 3897-3908. doi: 10.1093/nar/gkl466.
- Abstract
- The E2 proteins are transcription/replication factors from papillomaviruses. Human papillomaviruses (HPVs) can be broadly divided in two groups; low-risk HPV subtypes cause benign warts while high-risk HPVs give rise to cervical cancer. Although a range of crystal structures of E2 DNA-binding domains (DBD) from both high- and low-risk HPV subtypes have been reported previously, structures of E2 DBD:DNA complexes have only been available for high-risk HPV18 and bovine papillomavirus (BPV1). In the present study we report the unliganded and DNA complex structures of the E2 DBD from the low-risk HPV6. As in the previous E2-DNA structures, complex formation results in considerable bending of the DNA, which is facilitated by sequences with A:T-rich spacers that adopt a pre-bent conformation. The low-risk HPV6 E2-DNA complex differs from the earlier structures in that minimal deformation of the protein accompanies complex formation. Stopped-flow kinetic studies confirm that both high- and low-risk E2 proteins adapt their structures on binding to DNA, although this is achieved more readily for HPV6 E2. It therefore appears that the higher selectivity of the HPV6 E2 protein may arise from its limited molecular adaptability, a property that might distinguish the behaviour of E2 proteins from high- and low-risk HPV subtypes.