Summary information and primary citation
- PDB-id
- 2bh2; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transferase
- Method
- X-ray (2.15 Å)
- Summary
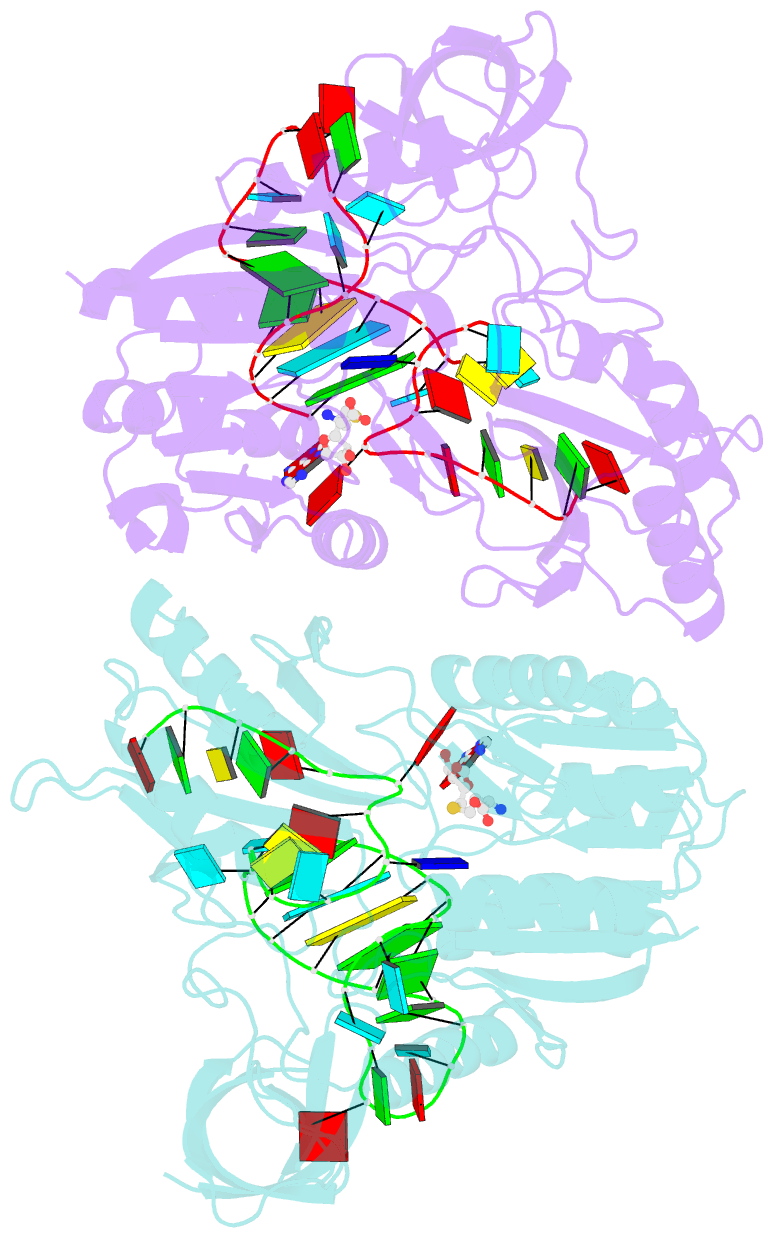
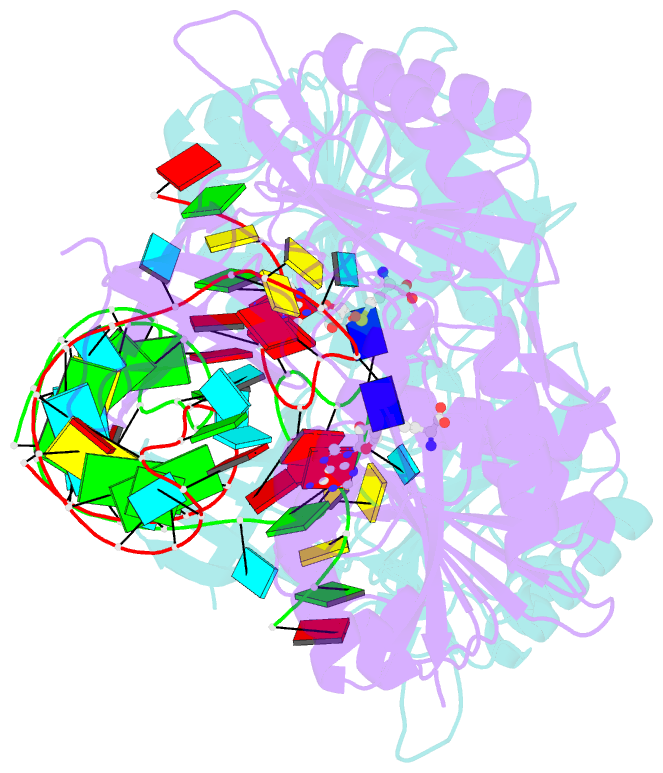
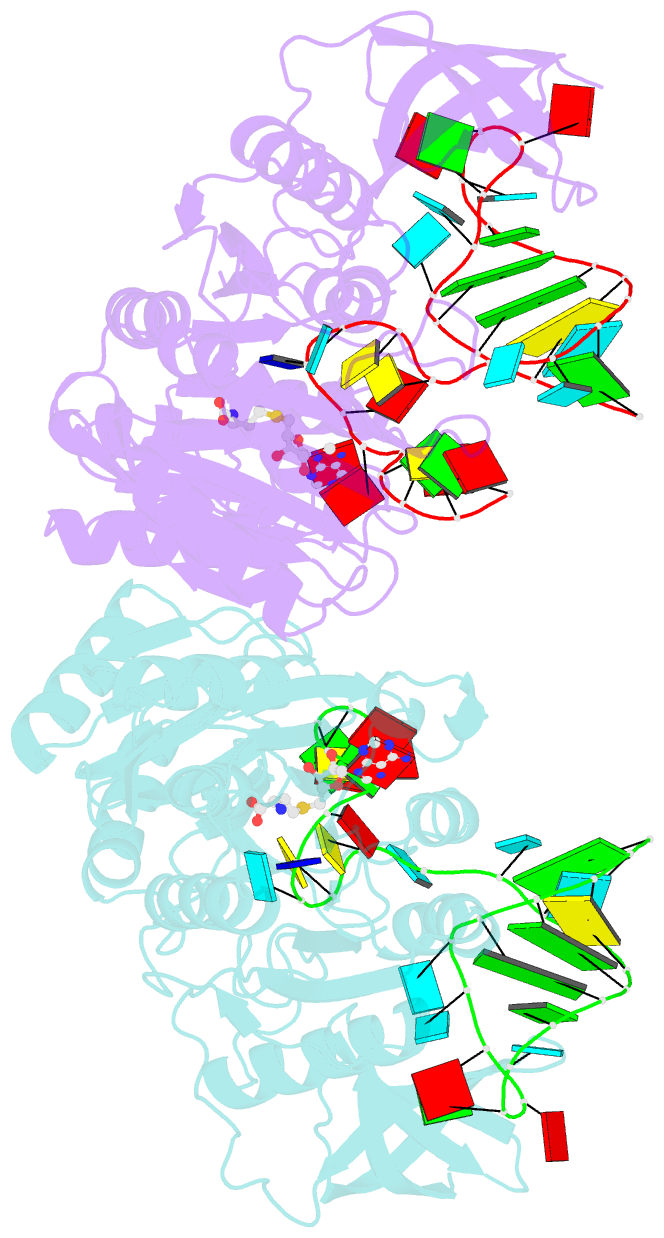
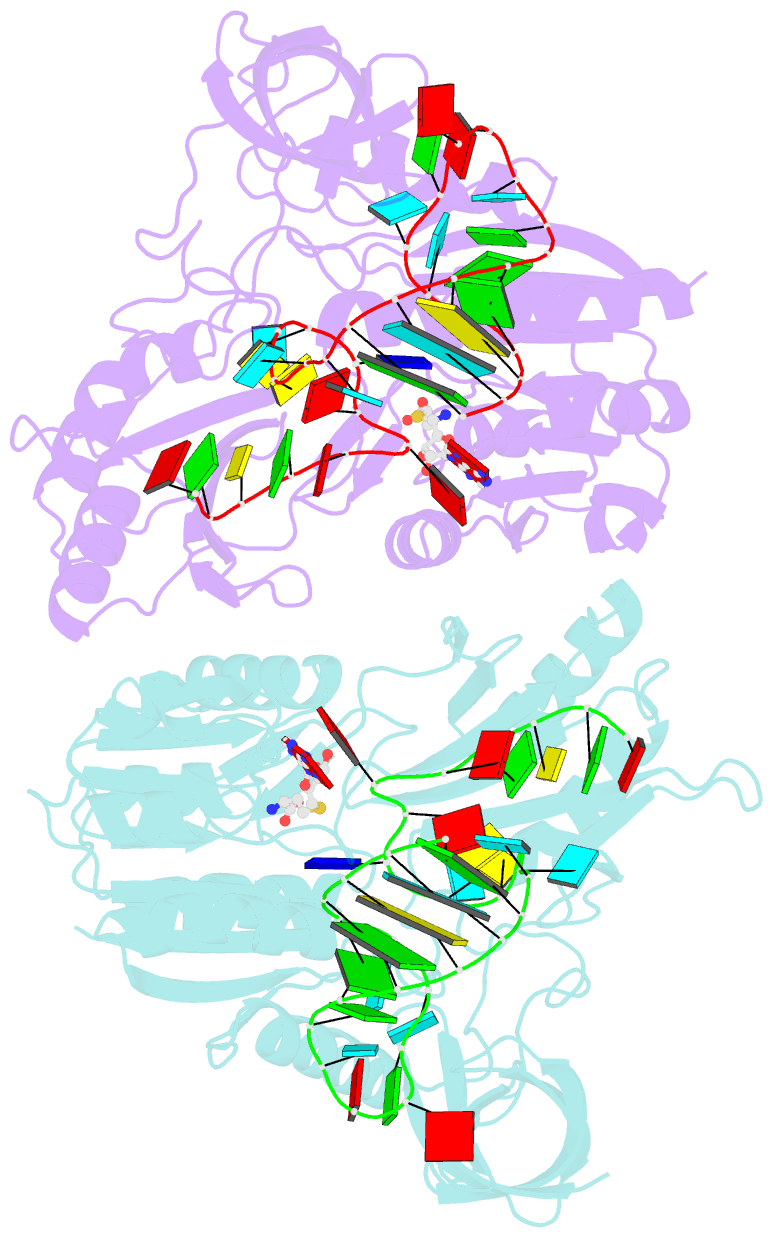
- Crystal structure of e. coli 5-methyluridine methyltransferase ruma in complex with ribosomal RNA substrate and s-adenosylhomocysteine.
- Reference
- Lee TT, Agarwalla S, Stroud RM (2005): "A Unique RNA Fold in the Ruma-RNA-Cofactor Ternary Complex Contributes to Substrate Selectivity and Enzymatic Function." Cell(Cambridge,Mass.), 120, 599. doi: 10.1016/J.CELL.2004.12.037.
- Abstract
- A single base (U1939) within E. coli 23S ribosomal RNA is methylated by its dedicated enzyme, RumA. The structure of RumA/RNA/S-adenosylhomocysteine uncovers the mechanism for achieving unique selectivity. The single-stranded substrate is "refolded" on the enzyme into a compact conformation with six key intra-RNA interactions. The RNA substrate contributes directly to catalysis. In addition to the target base, a second base is "flipped out" from the core loop to stack against the adenine of the cofactor S-adenosylhomocysteine. Nucleotides in permuted sequence order are stacked into the site vacated by the everted target U1939 and compensate for the energetic penalty of base eversion. The 3' hairpin segment of the RNA binds distal to the active site and provides binding energy that contributes to enhanced catalytic efficiency. Active collaboration of RNA in catalysis leads us to conclude that RumA and its substrate RNA may reflect features from the earliest RNA-protein era.