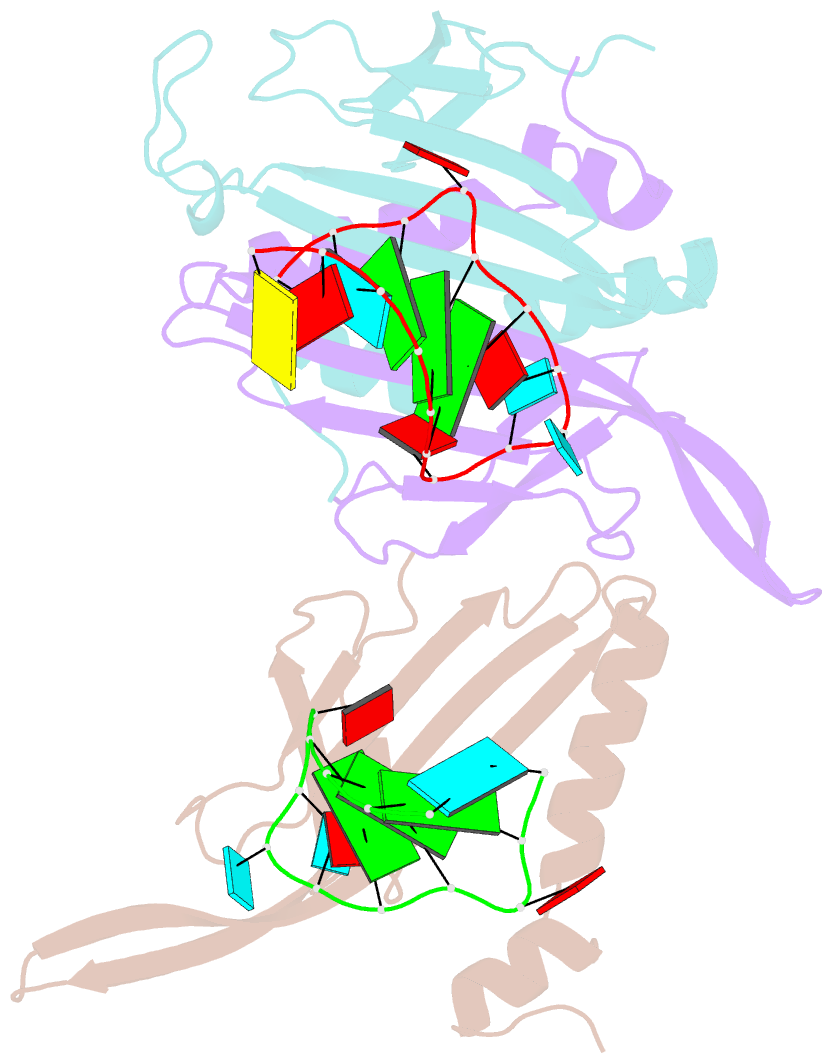
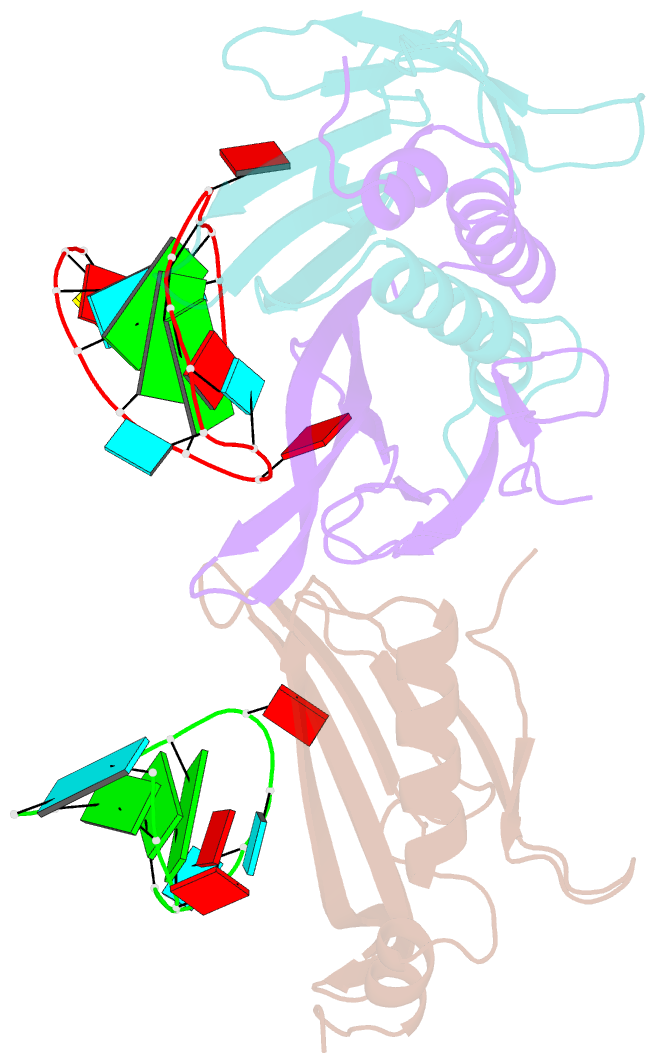
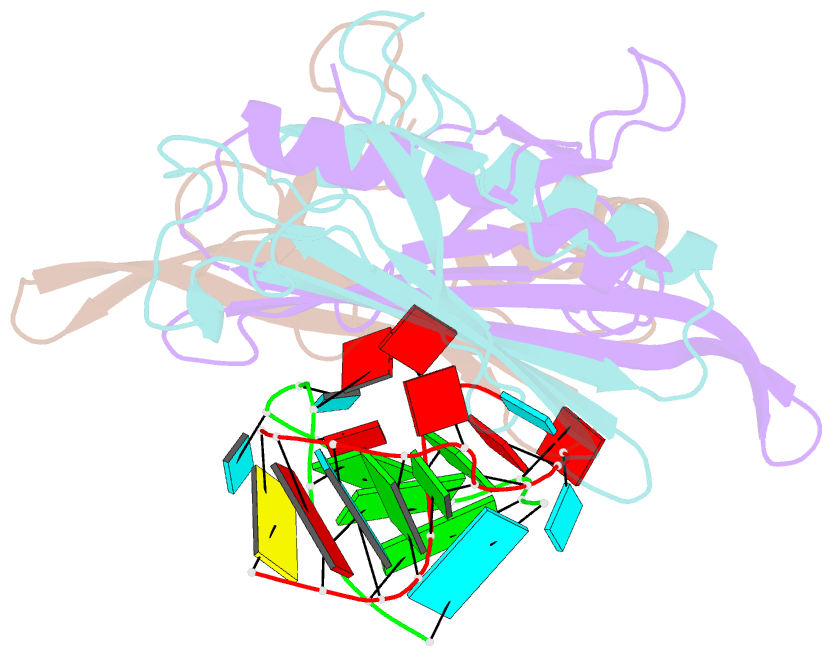
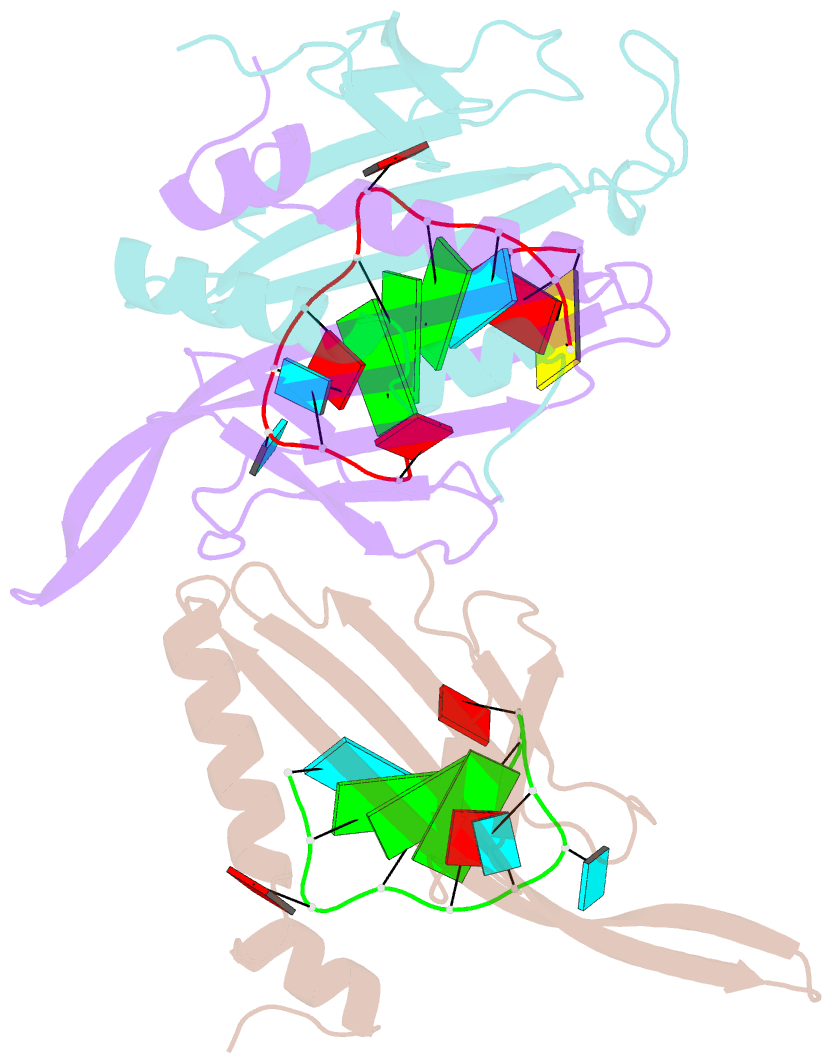
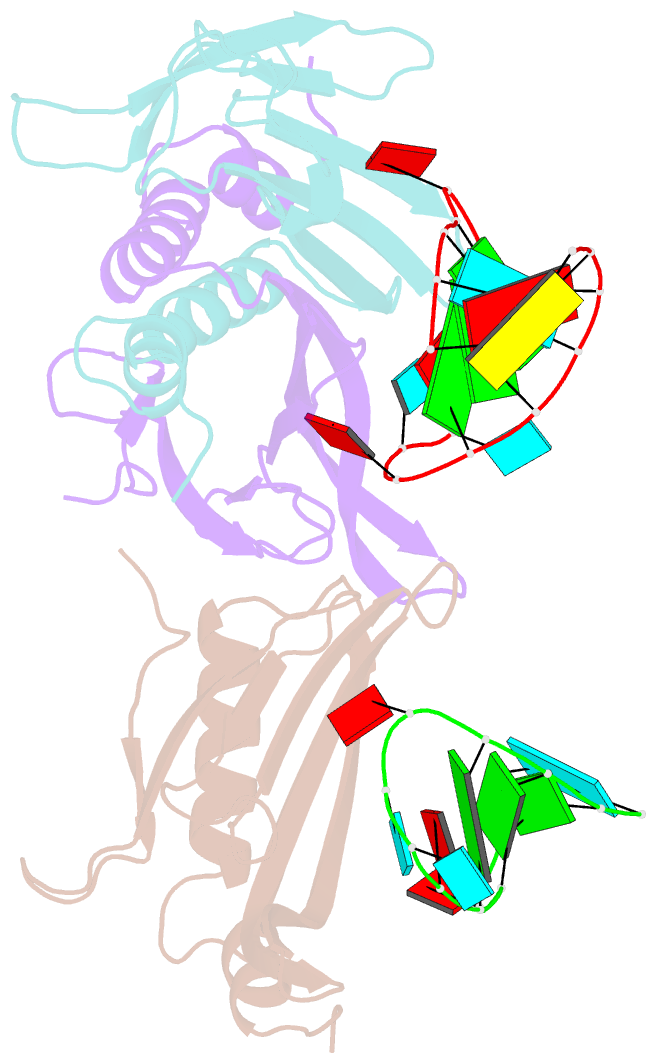
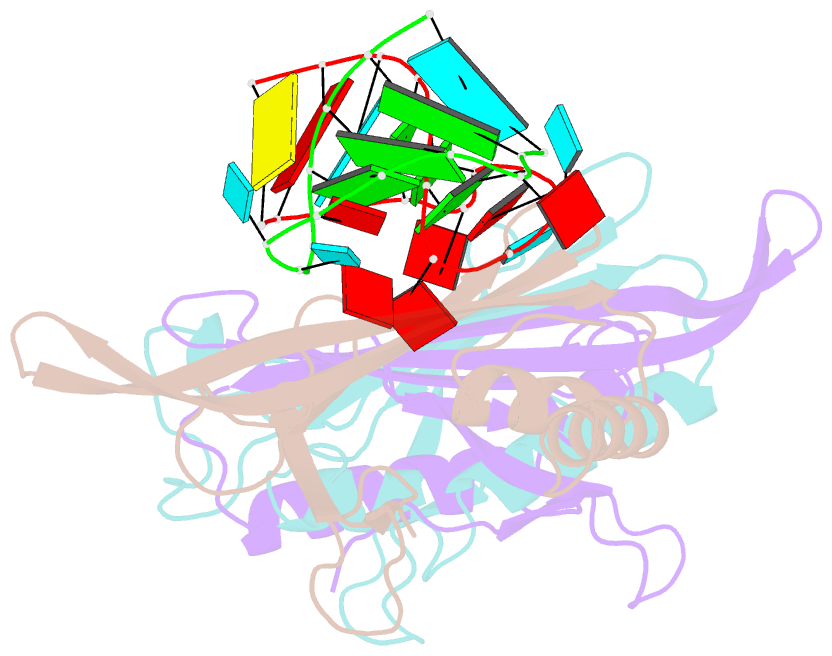
Summary information and primary citation
- PDB-id
- 2c4z; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- virus-RNA
- Method
- X-ray (2.6 Å)
- Summary
- Ms2-RNA hairpin (2su -5-6) complex
- Reference
- Grahn E, Moss T, Helgstrand C, Fridborg K, Sundaram M, Tars K, Lago H, Stonehouse NJ, Davis DR, Stockley PG, Liljas L (2001): "Structural basis of pyrimidine specificity in the MS2 RNA hairpin-coat-protein complex." Rna, 7, 1616-1627.
- Abstract
- We have determined the X-ray structures of six MS2 RNA hairpin-coat-protein complexes having five different substitutions at the hairpin loop base -5. This is a uracil in the wild-type hairpin and contacts the coat protein both by stacking on to a tyrosine side chain and by hydrogen bonding to an asparagine side chain. The RNA consensus sequence derived from coat protein binding studies with natural sequence variants suggested that the -5 base needs to be a pyrimidine for strong binding. The five -5 substituents used in this study were 5-bromouracil, pyrimidin-2-one, 2-thiouracil, adenine, and guanine. The structure of the 5-bromouracil complex was determined to 2.2 A resolution, which is the highest to date for any MS2 RNA-protein complex. All the complexes presented here show very similar conformations, despite variation in affinity in solution. The results suggest that the stacking of the -5 base on to the tyrosine side chain is the most important driving force for complex formation. A number of hydrogen bonds that are present in the wild-type complex are not crucial for binding, as they are missing in one or more of the complexes. The results also reveal the flexibility of this RNA-protein interface, with respect to functional group variation, and may be generally applicable to other RNA-protein complexes.