Summary information and primary citation
- PDB-id
- 2ct8; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- ligase-RNA
- Method
- X-ray (2.7 Å)
- Summary
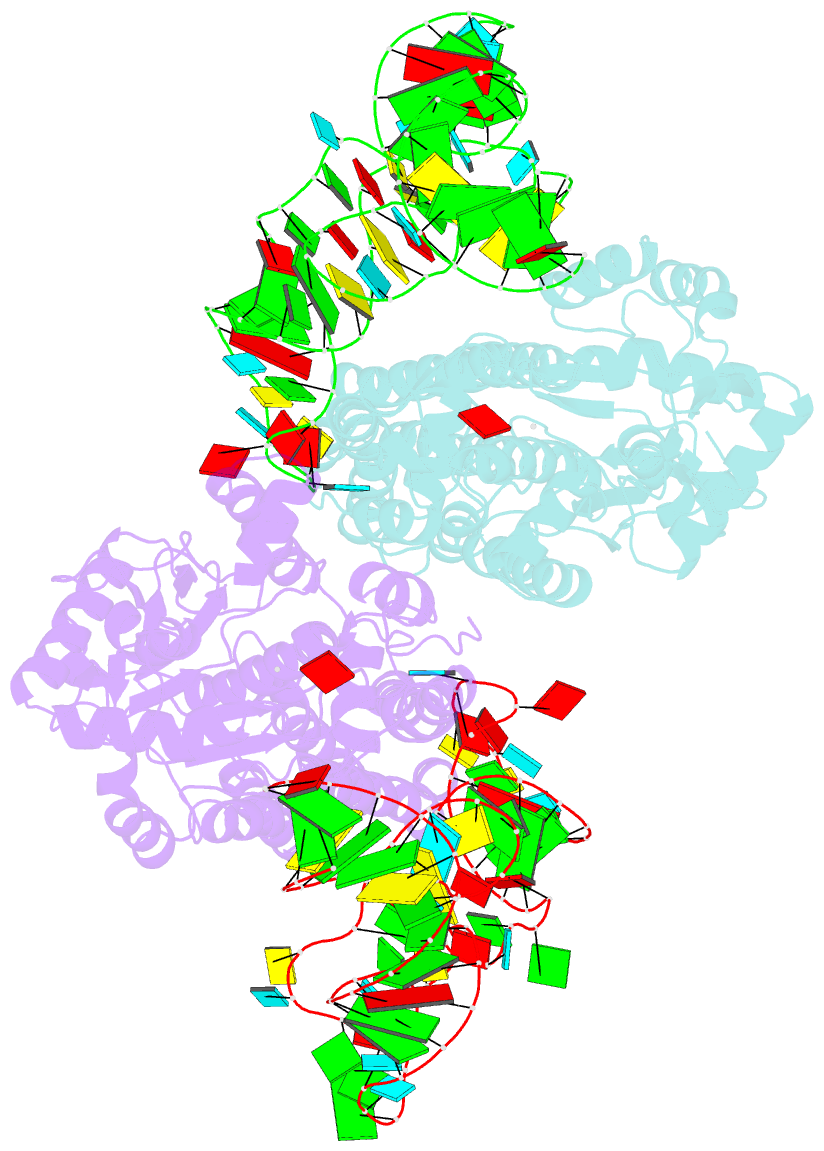
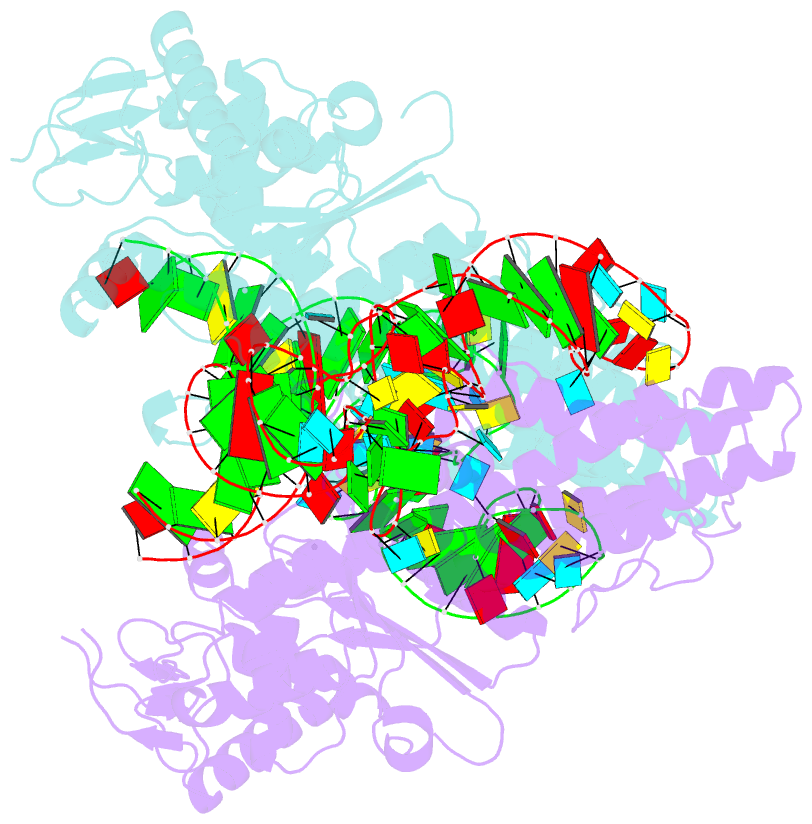
- Crystal structure of aquifex aeolicus methionyl-trna synthetase complexed with trna(met) and methionyl-adenylate anologue
- Reference
- Nakanishi K, Ogiso Y, Nakama T, Fukai S, Nureki O (2005): "Structural basis for anticodon recognition by methionyl-tRNA synthetase." Nat.Struct.Mol.Biol., 12, 931-932. doi: 10.1038/nsmb988.
- Abstract
- In the 2.7-A resolution crystal structure of methionyl-tRNA synthetase (MetRS) in complex with tRNA(Met) and a methionyl-adenylate analog, the tRNA anticodon loop is distorted to form a triple-base stack comprising C34, A35 and A38. A tryptophan residue stacks on C34 to extend the triple-base stack. In addition, C34 forms Watson-Crick-type hydrogen bonds with Arg357. This structure resolves the longstanding question of how MetRS specifically recognizes tRNA(Met).