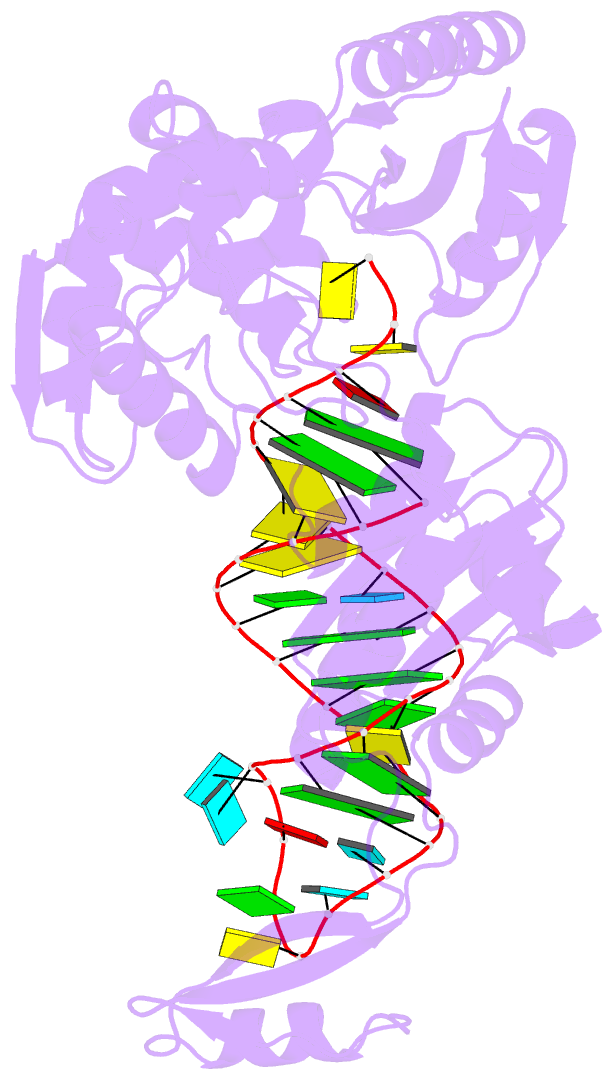
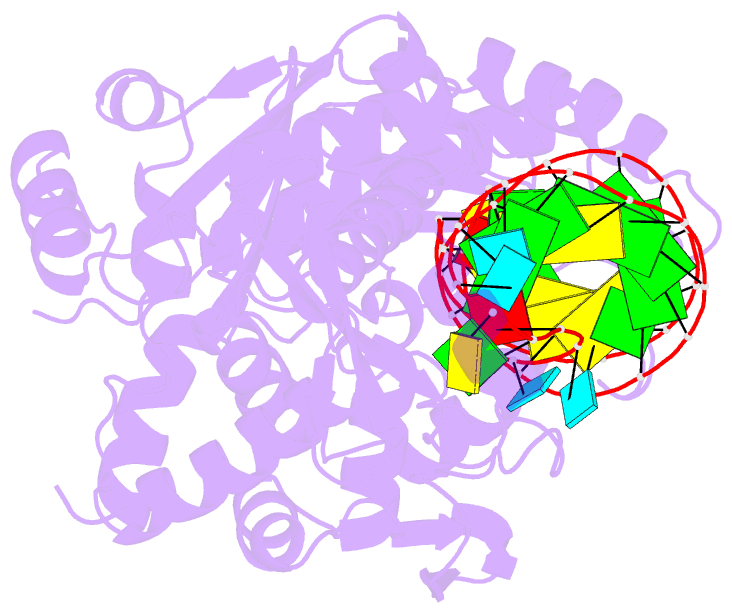
Summary information and primary citation
- PDB-id
- 2dr9; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transferase-RNA
- Method
- X-ray (2.8 Å)
- Summary
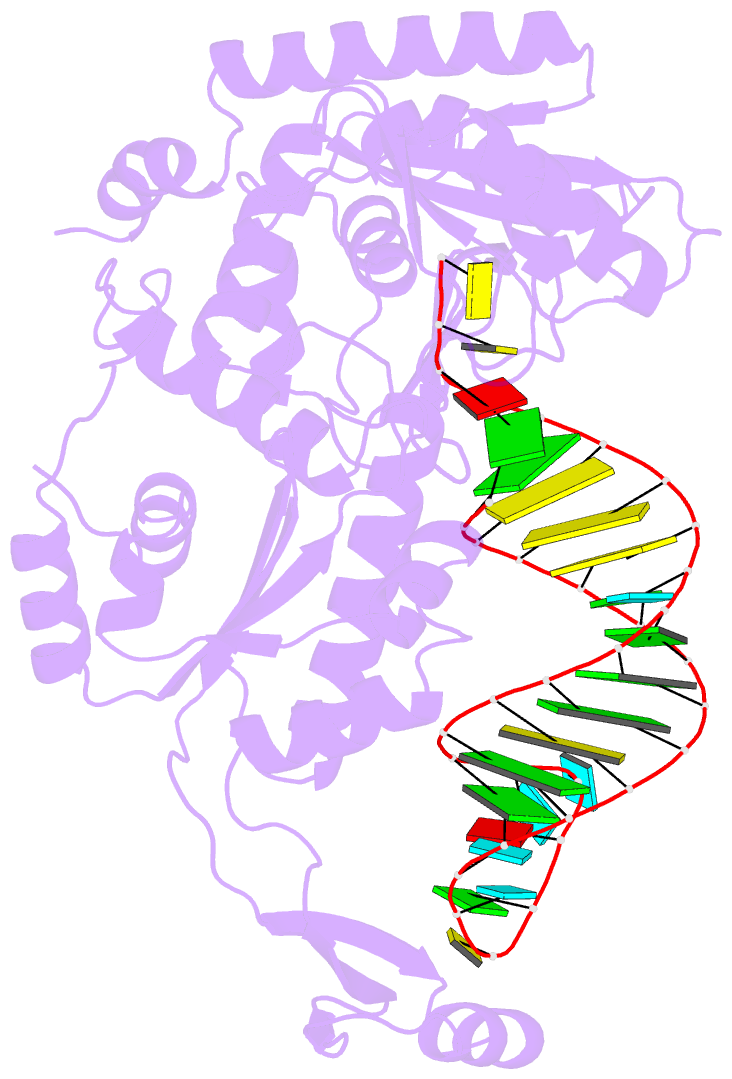
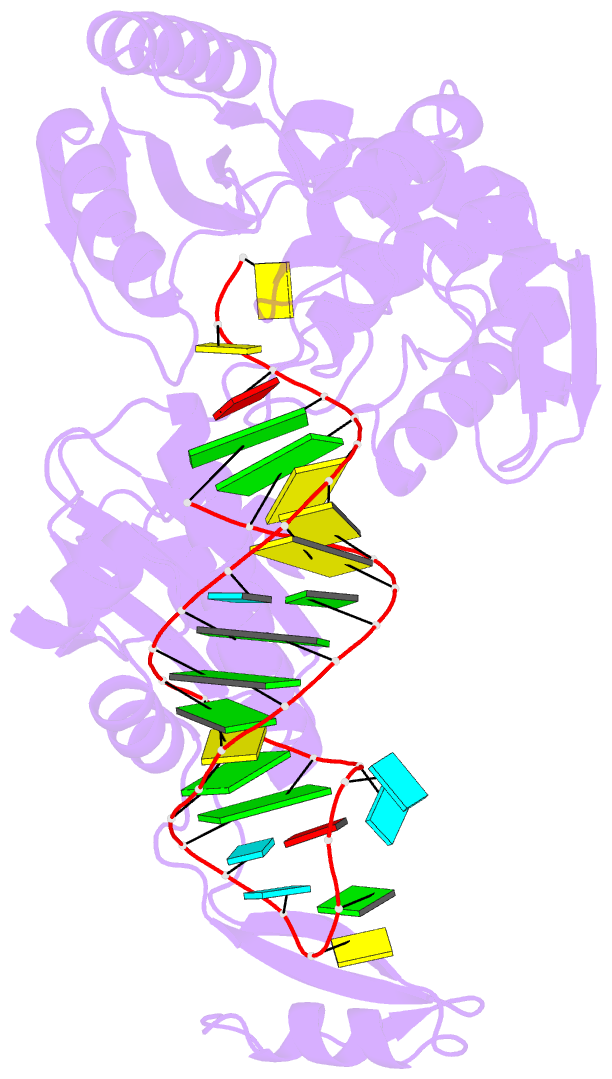
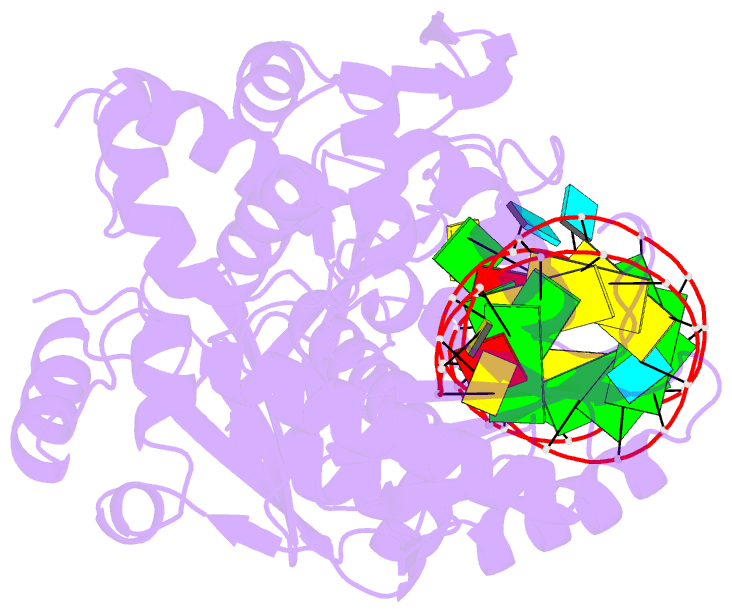
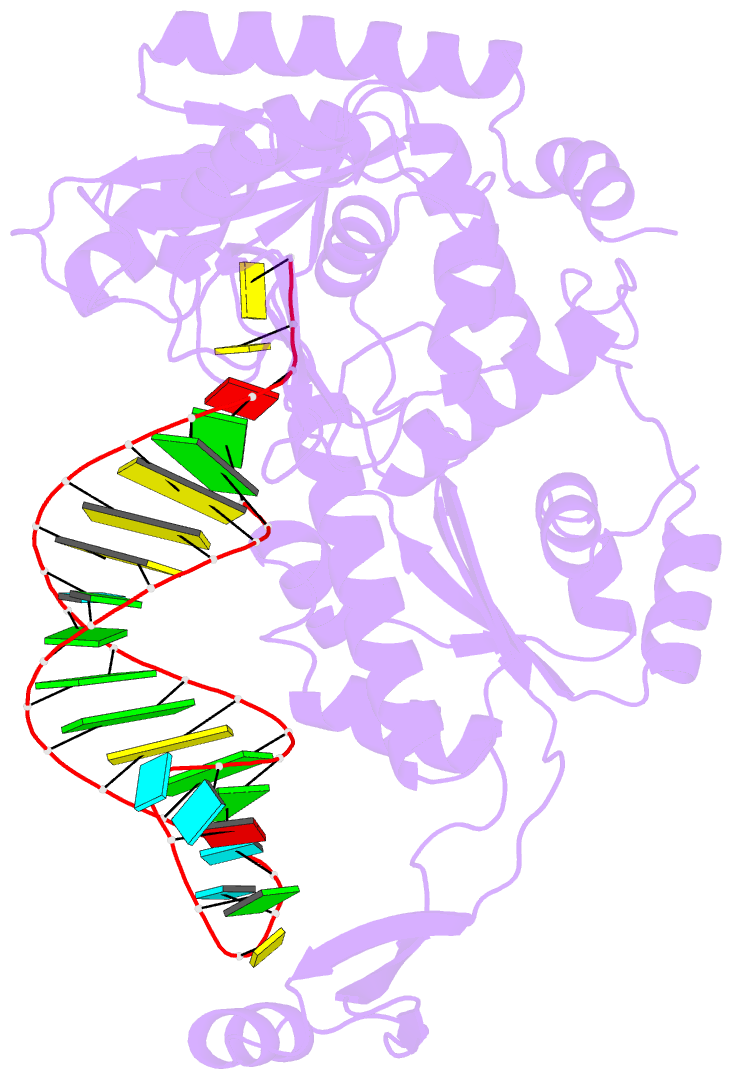
- Complex structure of cca-adding enzyme with trnaminidcc
- Reference
- Tomita K, Ishitani R, Fukai S, Nureki O (2006): "Complete crystallographic analysis of the dynamics of CCA sequence addition." Nature, 443, 956-960. doi: 10.1038/nature05204.
- Abstract
- CCA-adding polymerase matures the essential 3'-CCA terminus of transfer RNA without any nucleic-acid template. However, it remains unclear how the correct nucleotide triphosphate is selected in each reaction step and how the polymerization is driven by the protein and RNA dynamics. Here we present complete sequential snapshots of six complex structures of CCA-adding enzyme and four distinct RNA substrates with and without CTP (cytosine triphosphate) or ATP (adenosine triphosphate). The CCA-lacking RNA stem extends by one base pair to force the discriminator nucleoside into the active-site pocket, and then tracks back after incorporation of the first cytosine monophosphate (CMP). Accommodation of the second CTP clamps the catalytic cleft, inducing a reorientation of the turn, which flips C74 to allow CMP to be accepted. In contrast, after the second CMP is added, the polymerase and RNA primer are locked in the closed state, which directs the subsequent A addition. Between the CTP- and ATP-binding stages, the side-chain conformation of Arg 224 changes markedly; this is controlled by the global motion of the enzyme and position of the primer terminus, and is likely to achieve the CTP/ATP discrimination, depending on the polymerization stage. Throughout the CCA-adding reaction, the enzyme tail domain firmly anchors the TPsiC-loop of the tRNA, which ensures accurate polymerization and termination.