Summary information and primary citation
- PDB-id
- 2drp; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transcription-DNA
- Method
- X-ray (2.8 Å)
- Summary
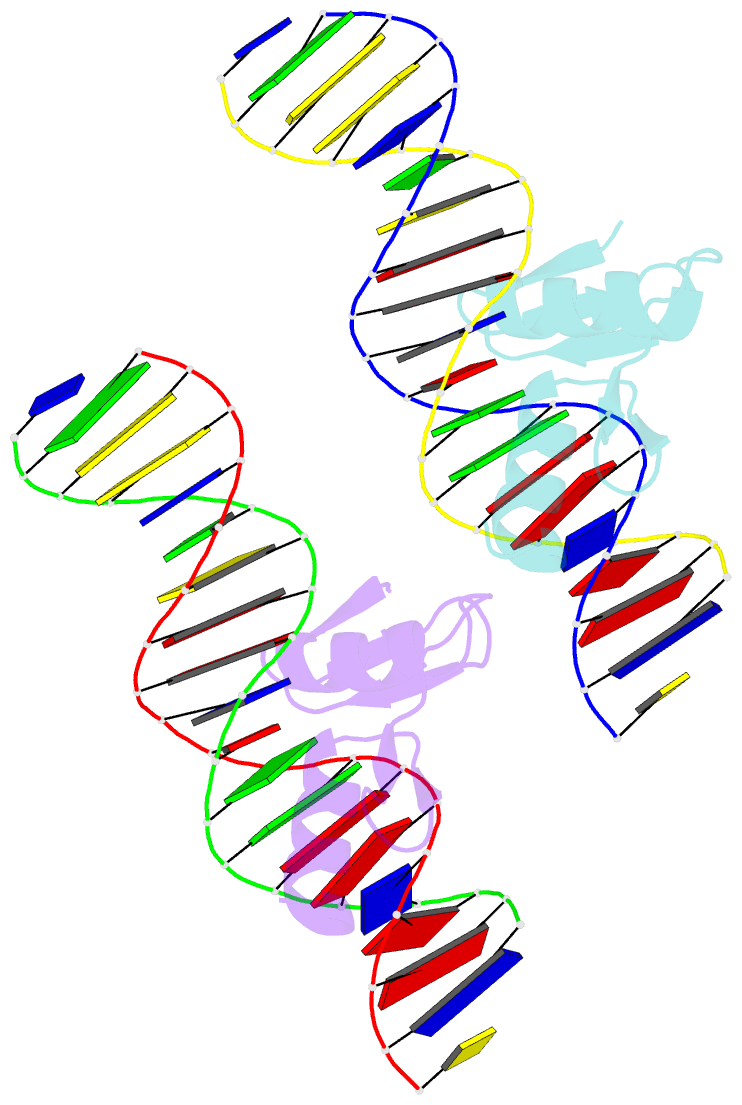
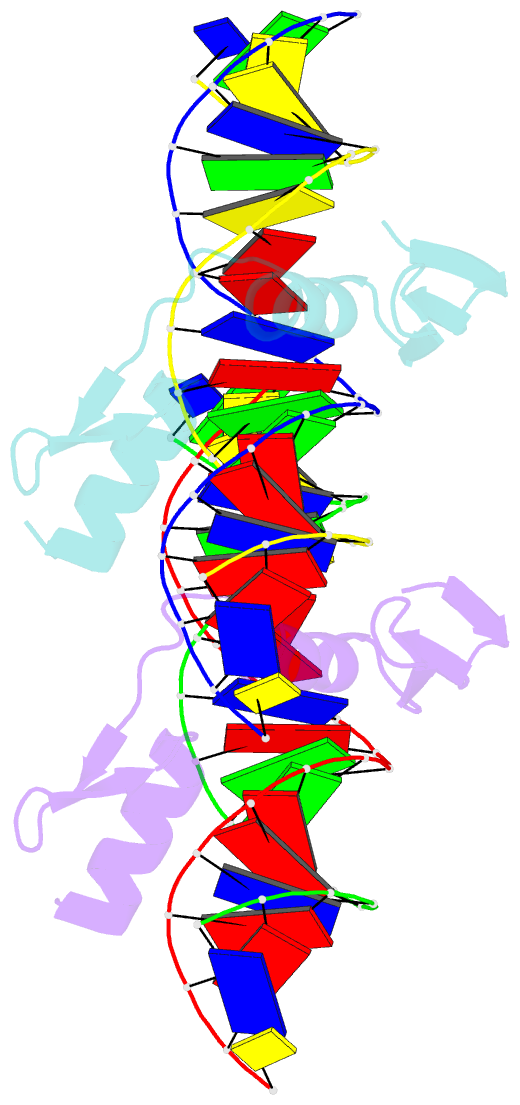
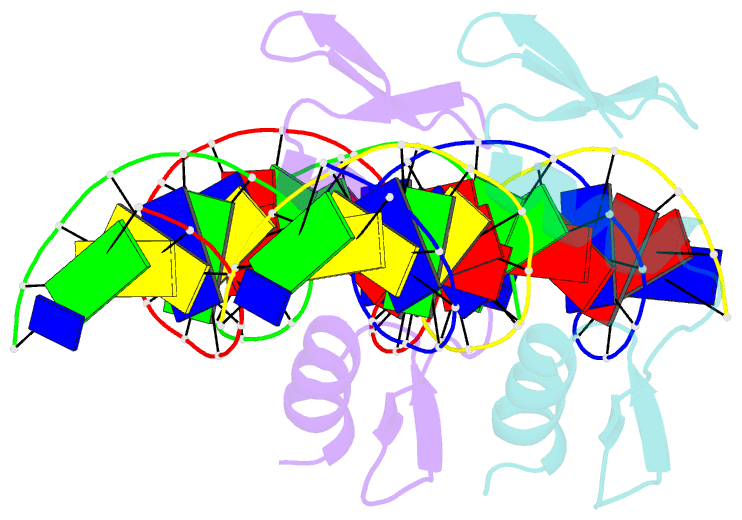
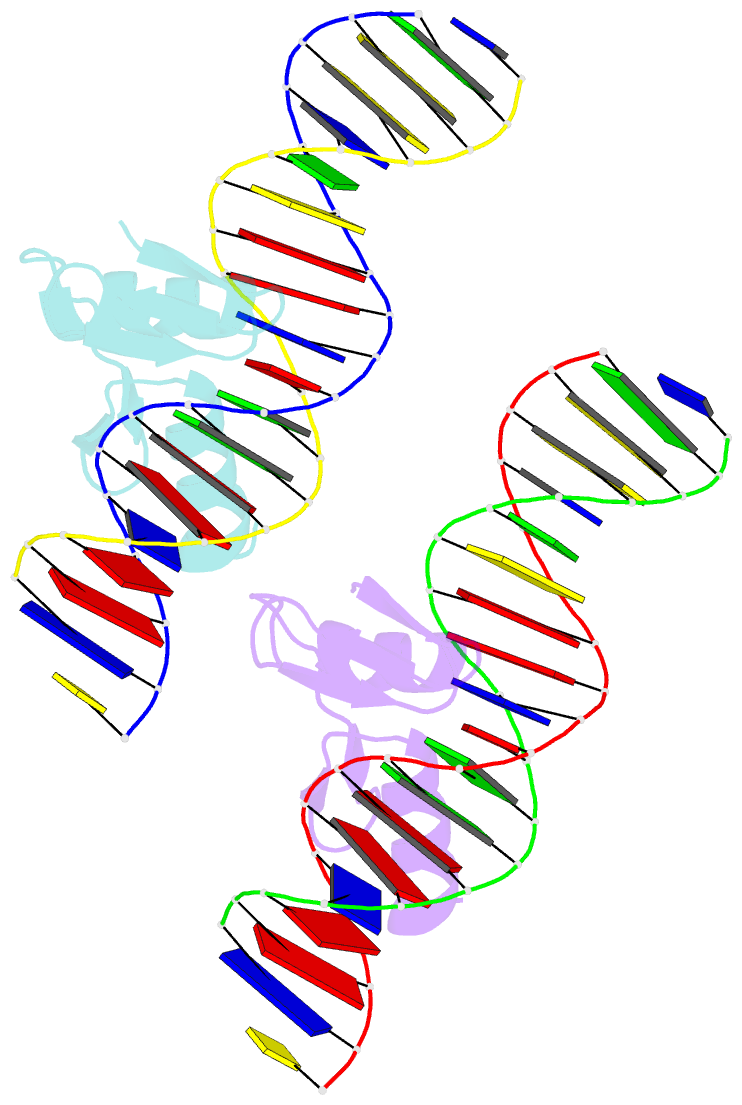
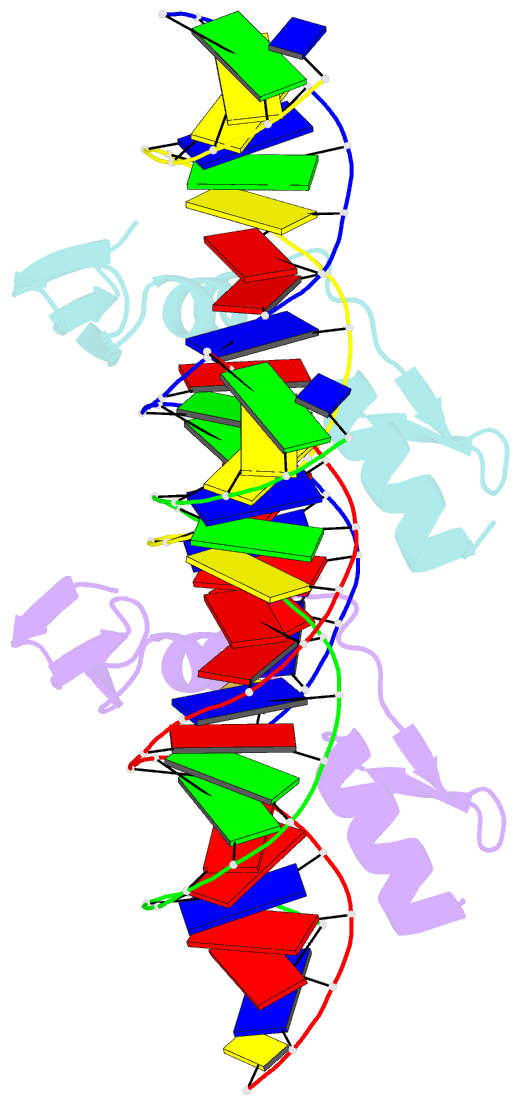
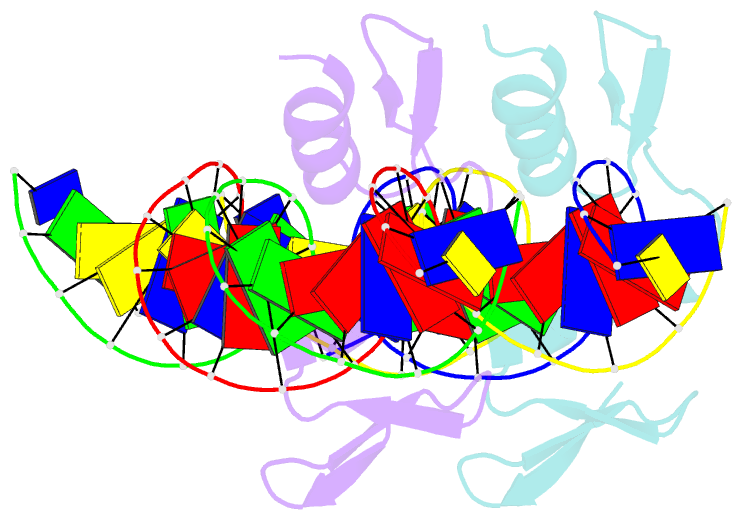
- The crystal structure of a two zinc-finger peptide reveals an extension to the rules for zinc-finger-DNA recognition
- Reference
- Fairall L, Schwabe JW, Chapman L, Finch JT, Rhodes D (1993): "The crystal structure of a two zinc-finger peptide reveals an extension to the rules for zinc-finger/DNA recognition." Nature, 366, 483-487. doi: 10.1038/366483a0.
- Abstract
- The Cys2-His2 zinc-finger is the most widely occurring DNA-binding motif. The first structure of a zinc-finger/DNA complex revealed a fairly simple mechanism for DNA recognition suggesting that the zinc-finger might represent a candidate template for designing proteins to recognize DNA. Residues at three key positions in an alpha-helical 'reading head' play a dominant role in base-recognition and have been targets for mutagenesis experiments aimed at deriving a recognition code. Here we report the structure of a two zinc-finger DNA-binding domain from the protein Tramtrack complexed with DNA. The amino-terminal zinc-finger and its interaction with DNA illustrate several novel features. These include the use of a serine residue, which is semi-conserved and located outside the three key positions, to make a base contact. Its role in base-recognition correlates with a large, local, protein-induced deformation of the DNA helix at a flexible A-T-A sequence and may give insight into previous mutagenesis experiments. It is apparent from this structure that zinc-finger/DNA recognition is more complex than was originally perceived.