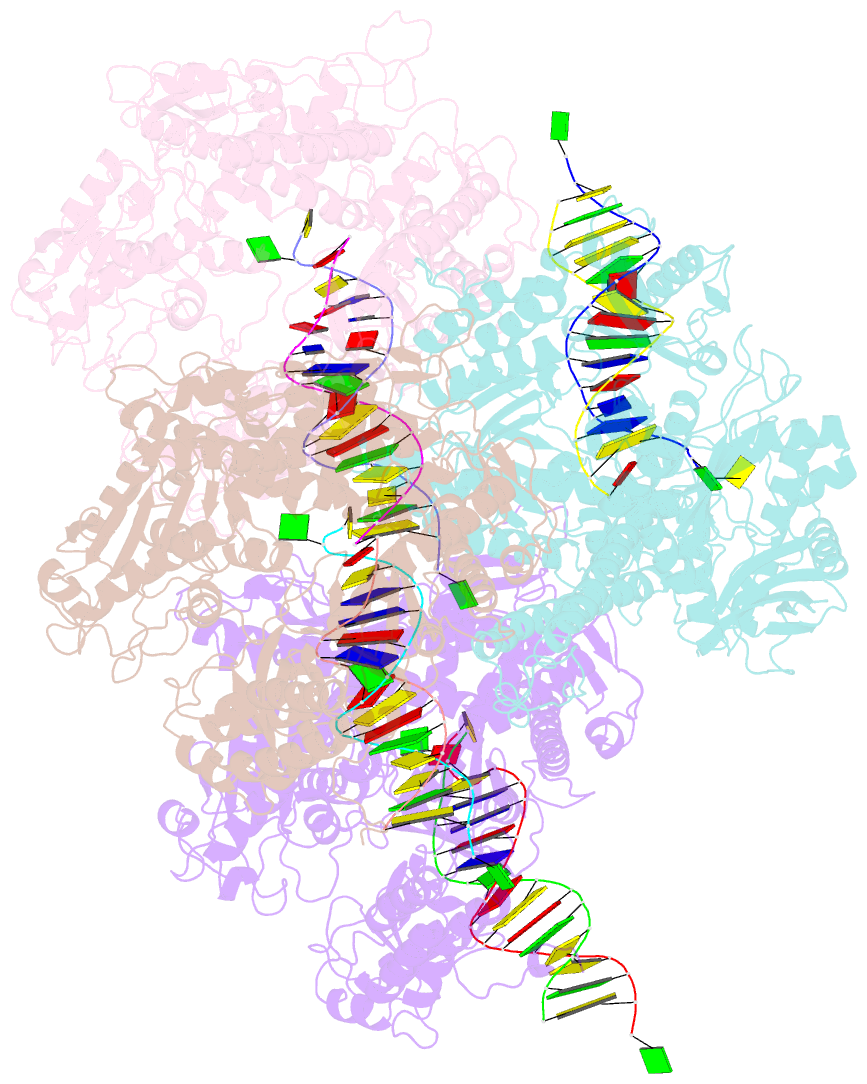
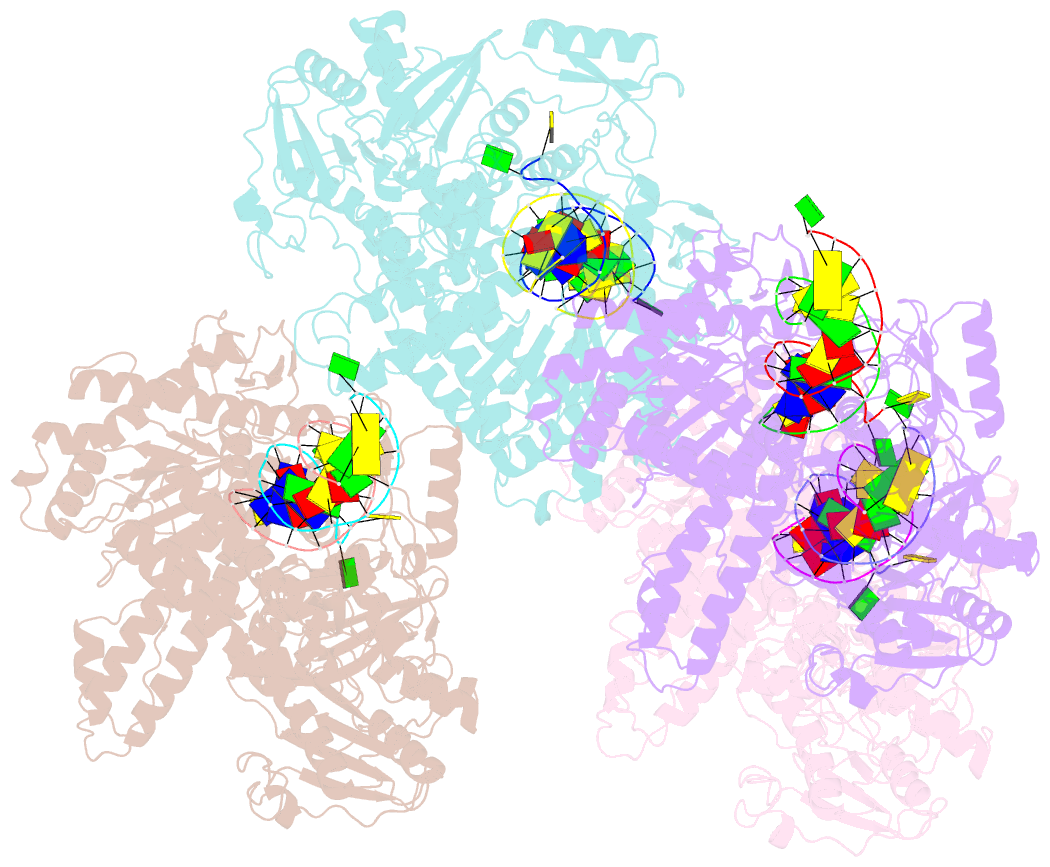
Summary information and primary citation
- PDB-id
- 2dtu; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transferase-DNA
- Method
- X-ray (2.37 Å)
- Summary
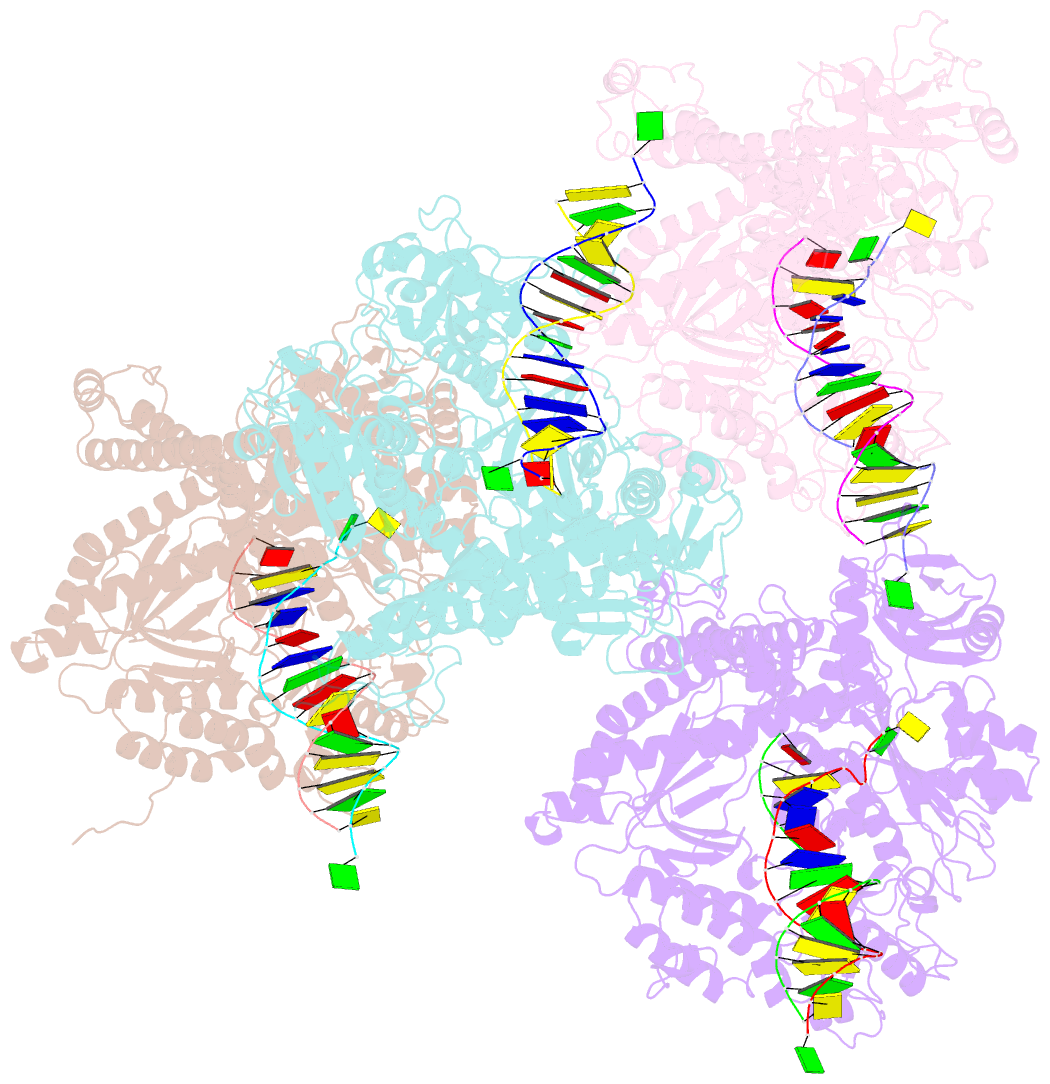
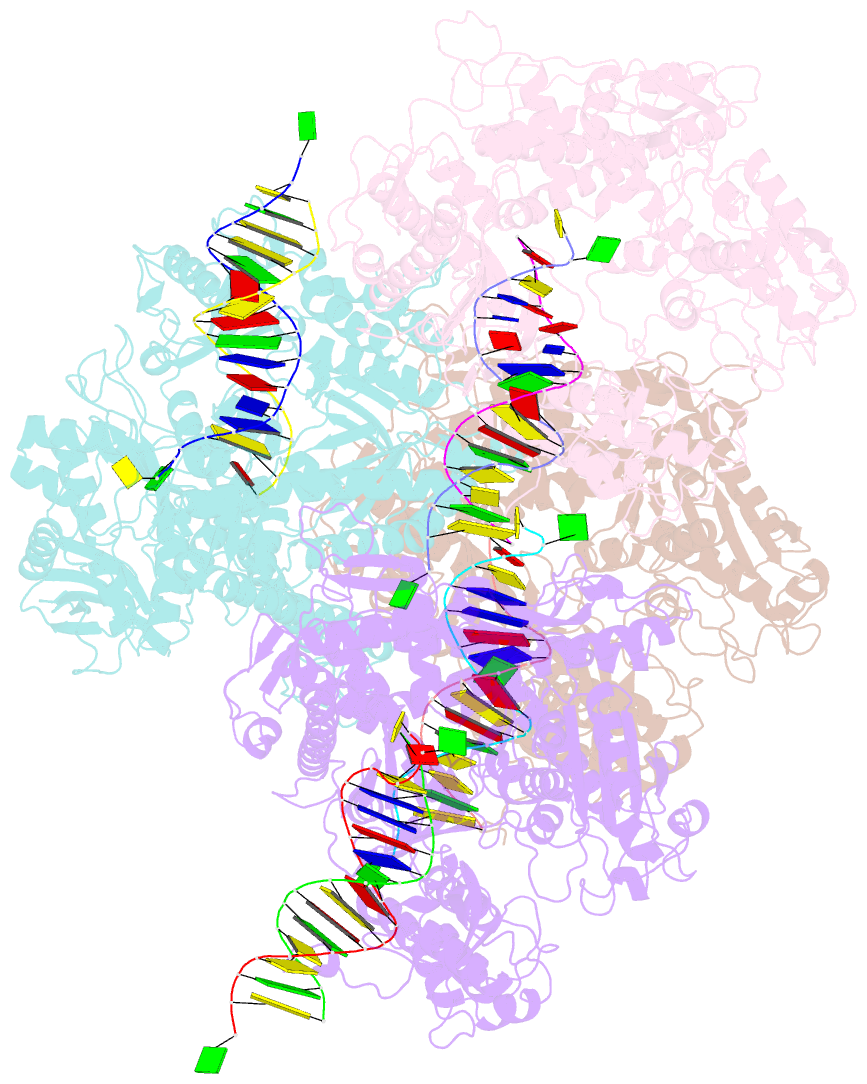
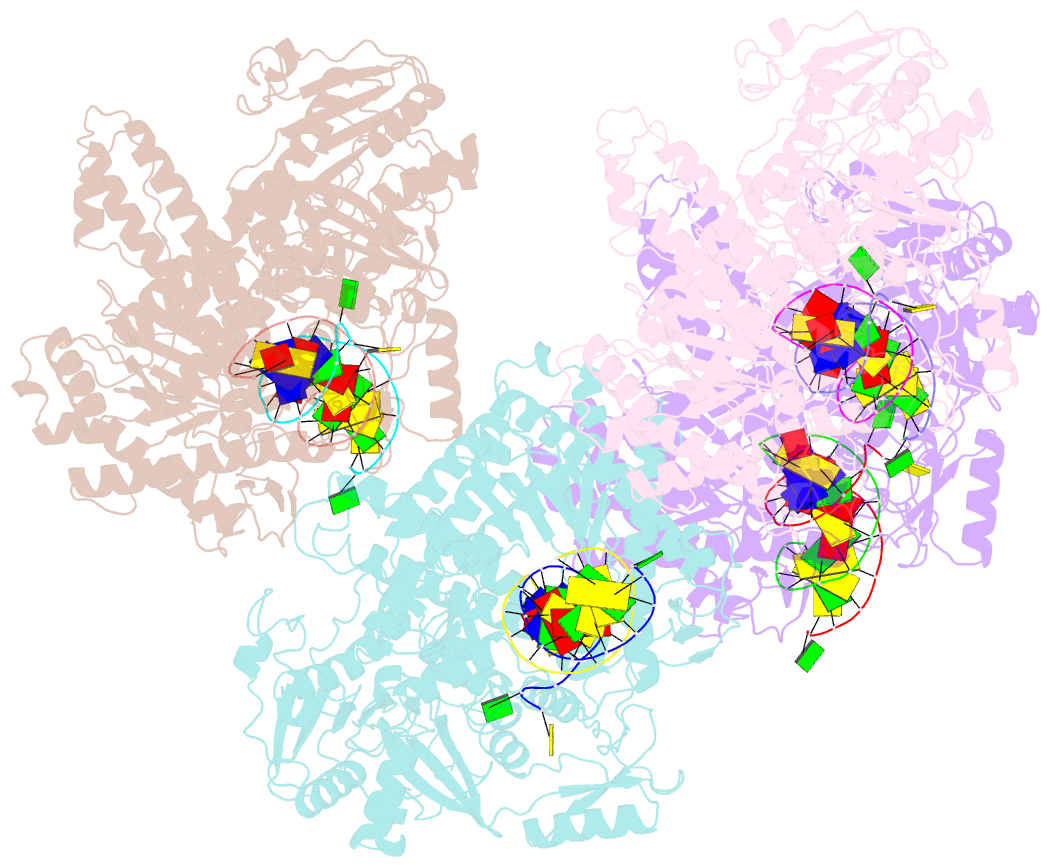
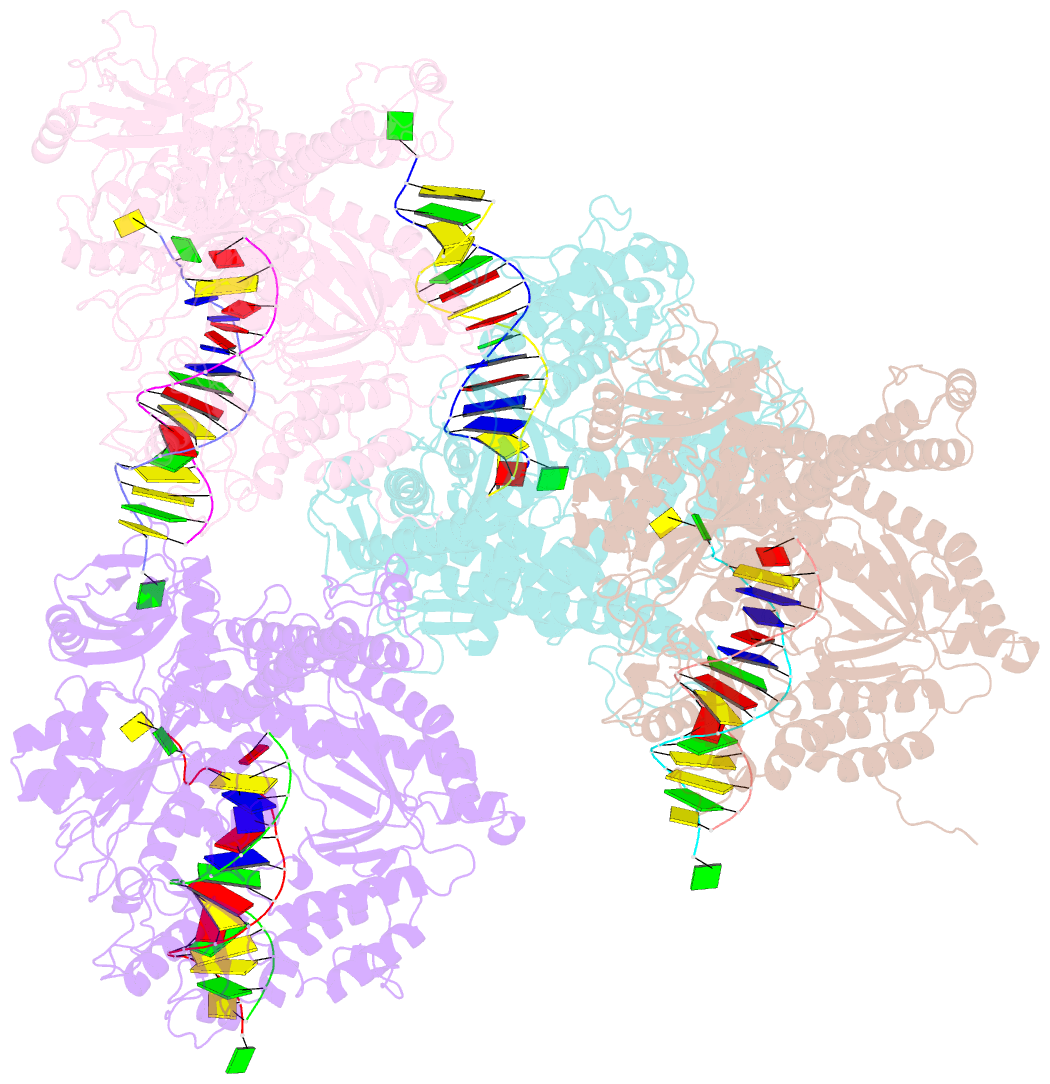
- Crystal structure of the beta hairpin loop deletion variant of rb69 gp43 in complex with DNA containing an abasic site analog
- Reference
- Hogg M, Aller P, Konigsberg W, Wallace SS, Doublie S (2007): "Structural and biochemical investigation of the role in proofreading of a beta hairpin loop found in the exonuclease domain of a replicative DNA polymerase of the B family." J.Biol.Chem., 282, 1432-1444. doi: 10.1074/jbc.M605675200.
- Abstract
- Replicative DNA polymerases, as exemplified by the B family polymerases from bacteriophages T4 and RB69, not only replicate DNA but also have the ability to proofread misincorporated nucleotides. Because the two activities reside in separate protein domains, polymerases must employ a mechanism that allows for efficient switching of the primer strand between the two active sites to achieve fast and accurate replication. Prior mutational and structural studies suggested that a beta hairpin structure located in the exonuclease domain of family B polymerases might play an important role in active site switching in the event of a nucleotide misincorporation. We show that deleting the beta hairpin loop in RB69 gp43 affects neither polymerase nor exonuclease activities. Single binding event studies with mismatched primer termini, however, show that the beta hairpin plays a role in maintaining the stability of the polymerase/DNA interactions during the binding of the primer DNA in the exonuclease active site but not on the return of the corrected primer to the polymerase active site. In addition, the deletion variant showed a more stable incorporation of a nucleotide opposite an abasic site. Moreover, in the 2.4 A crystal structure of the beta hairpin deletion variant incorporating an A opposite a templating furan, all four molecules in the crystal asymmetric unit have DNA in the polymerase active site, despite the presence of DNA distortions because of the misincorporation, confirming that the primer strand is not stably bound within the exonuclease active site in the absence of the beta hairpin loop.