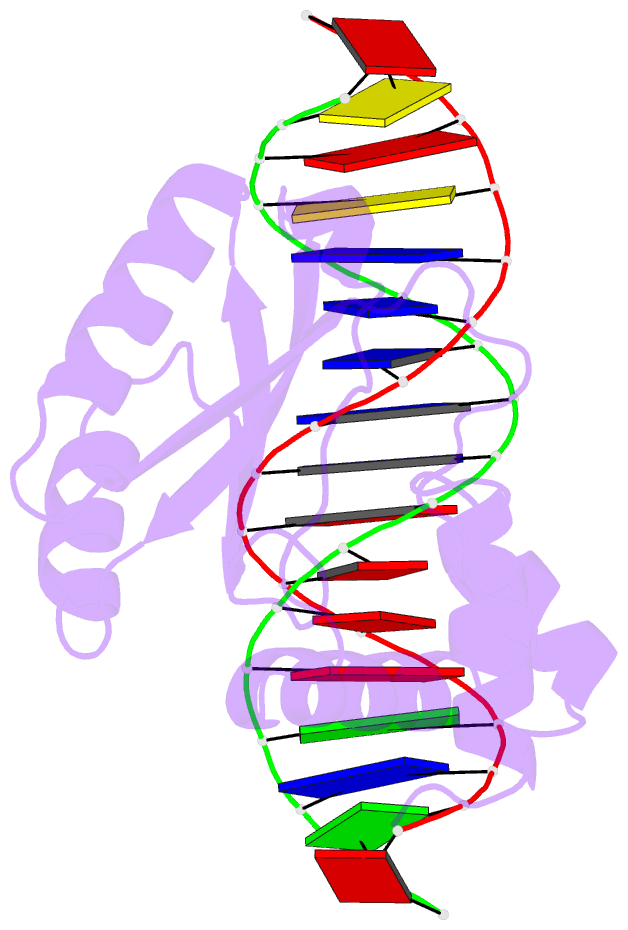
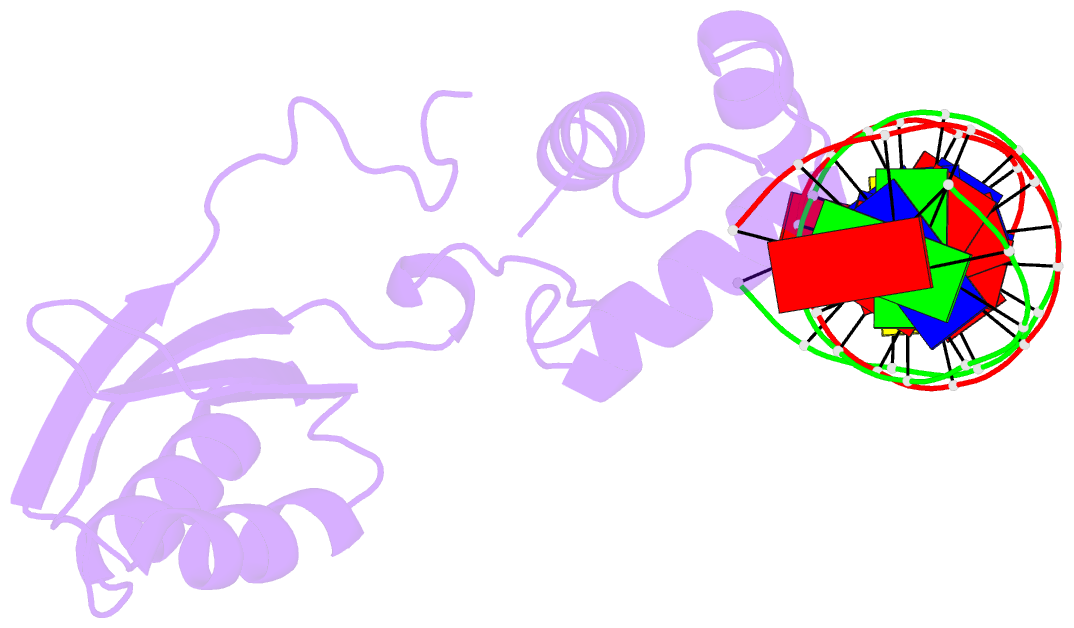
Summary information and primary citation
- PDB-id
- 2e1c; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transcription-DNA
- Method
- X-ray (2.1 Å)
- Summary
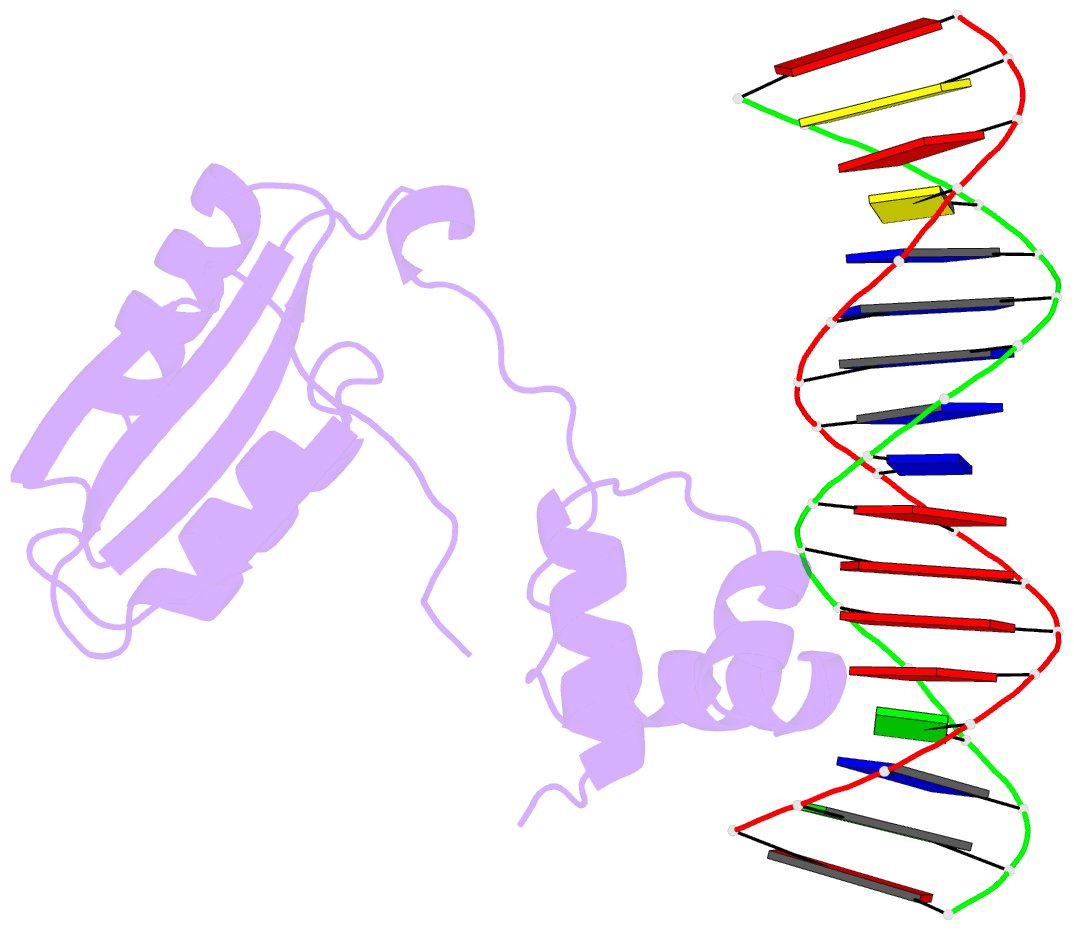
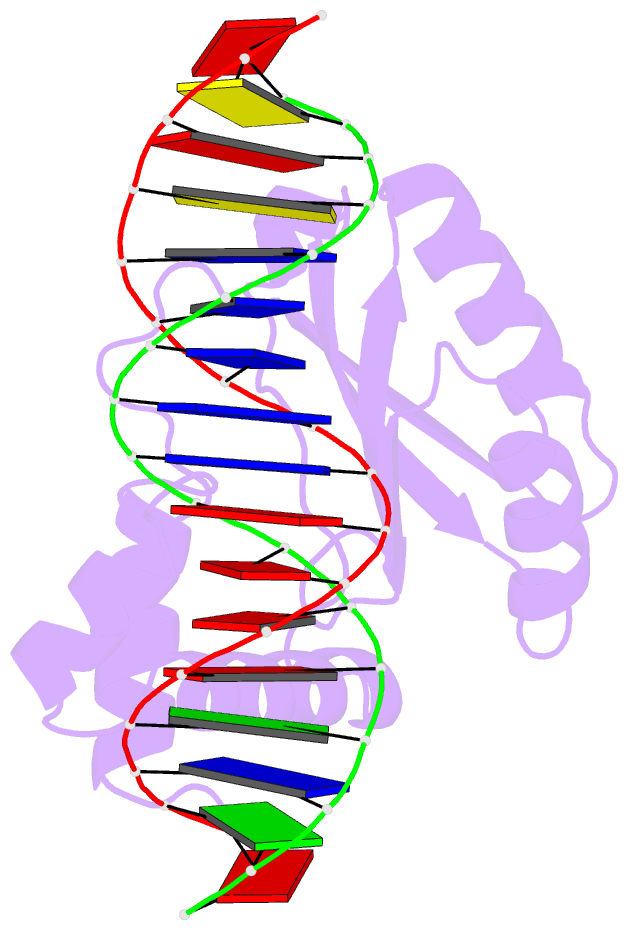
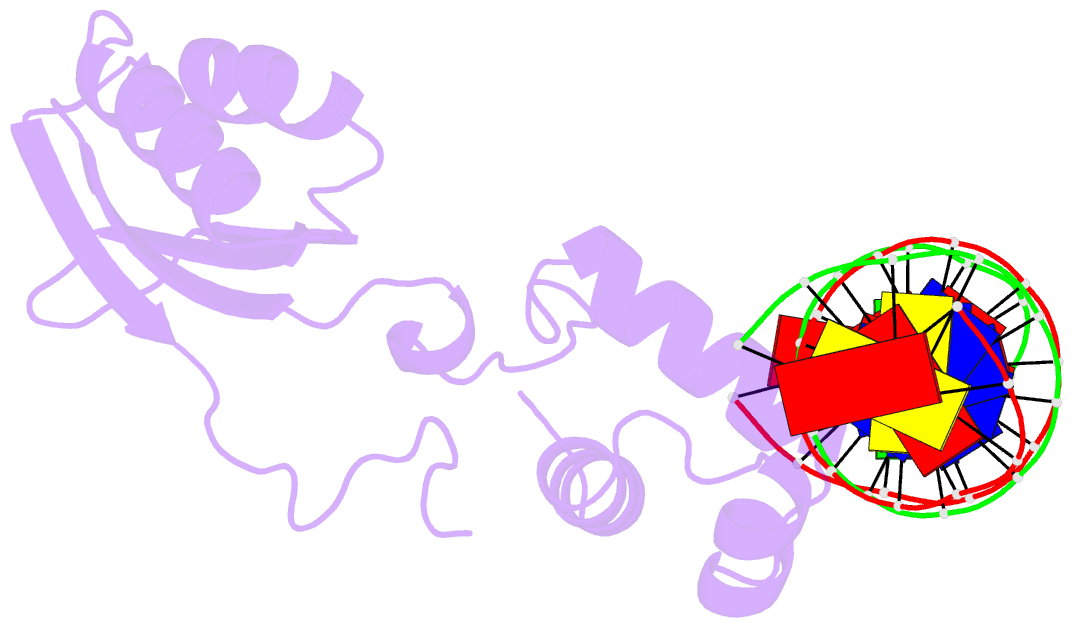
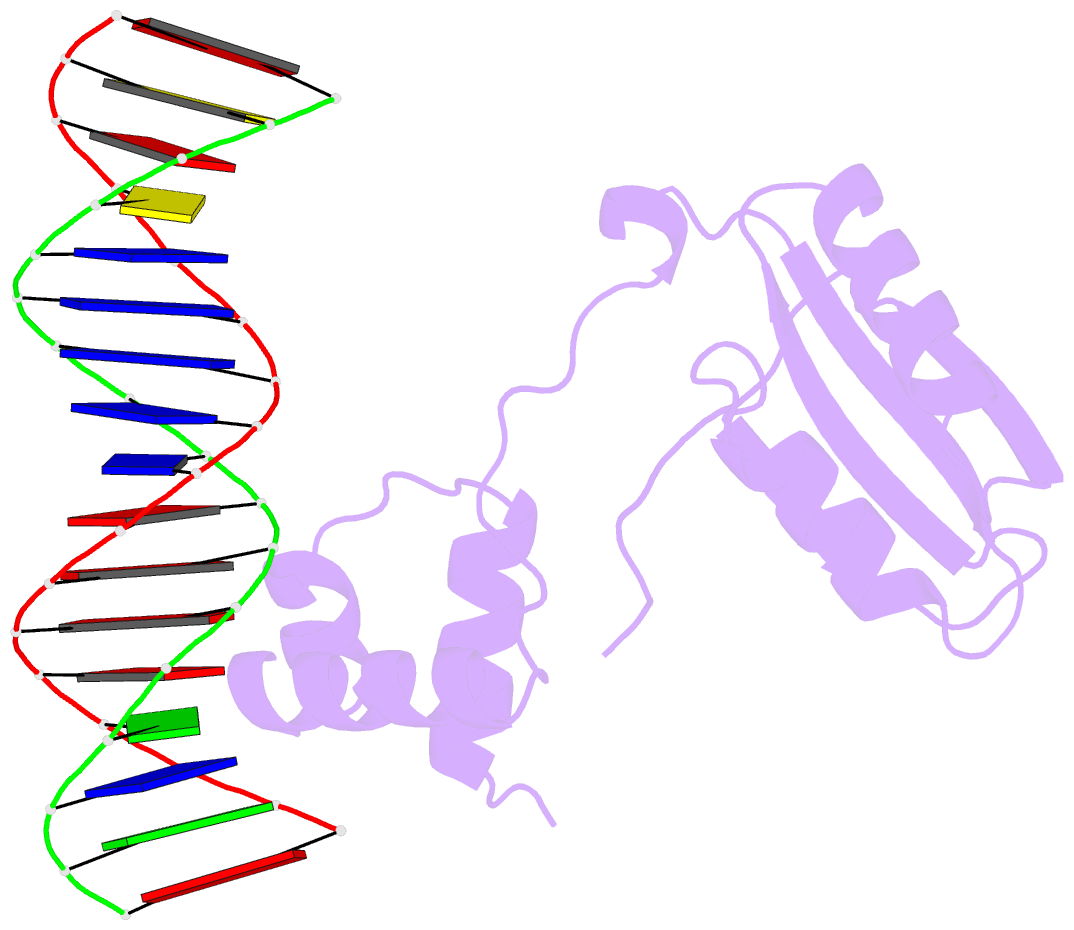
- Structure of putative hth-type transcriptional regulator ph1519-DNA complex
- Reference
- Yokoyama K, Ishijima SA, Koike H, Kurihara C, Shimowasa A, Kabasawa M, Kawashima T, Suzuki M (2007): "Feast/Famine Regulation by Transcription Factor FL11 for the Survival of the Hyperthermophilic Archaeon Pyrococcus OT3." Structure, 15, 1542-1554. doi: 10.1016/j.str.2007.10.015.
- Abstract
- Transcriptional repressor FL11 from the hyperthermophilic archaeon, Pyrococcus OT3, was crystallized in its dimer form in complex with a DNA duplex, TGAAAWWWTTTCA. Chemical contacting of FL11 to the terminal 5 bps, and DNA bending by propeller twisting at WWW confirmed specificity of the interaction. Dimer-binding sites were identified in promoters of approximately 200 transcription units coding, for example, H+-ATPase and NAD(P)H dehydrogenase. In the presence of lysine, four FL11 dimers were shown to assemble into an octamer, thereby covering the fl11 promoter. In the "feast" mode, when P. OT3 grows on amino acids, the FL11 octamer will terminate transcription of fl11, as was shown in vitro, thereby derepressing transcription of many metabolic genes. In the "famine" mode in the absence of lysine, approximately 6000 FL11 dimers present per cell will arrest growth. This regulation resembles global regulation by Escherichia coli leucine-responsive regulatory protein, and hints at a prototype of transcription regulations now highly diverged.