Summary information and primary citation
- PDB-id
- 2e52; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- hydrolase-DNA
- Method
- X-ray (2.0 Å)
- Summary
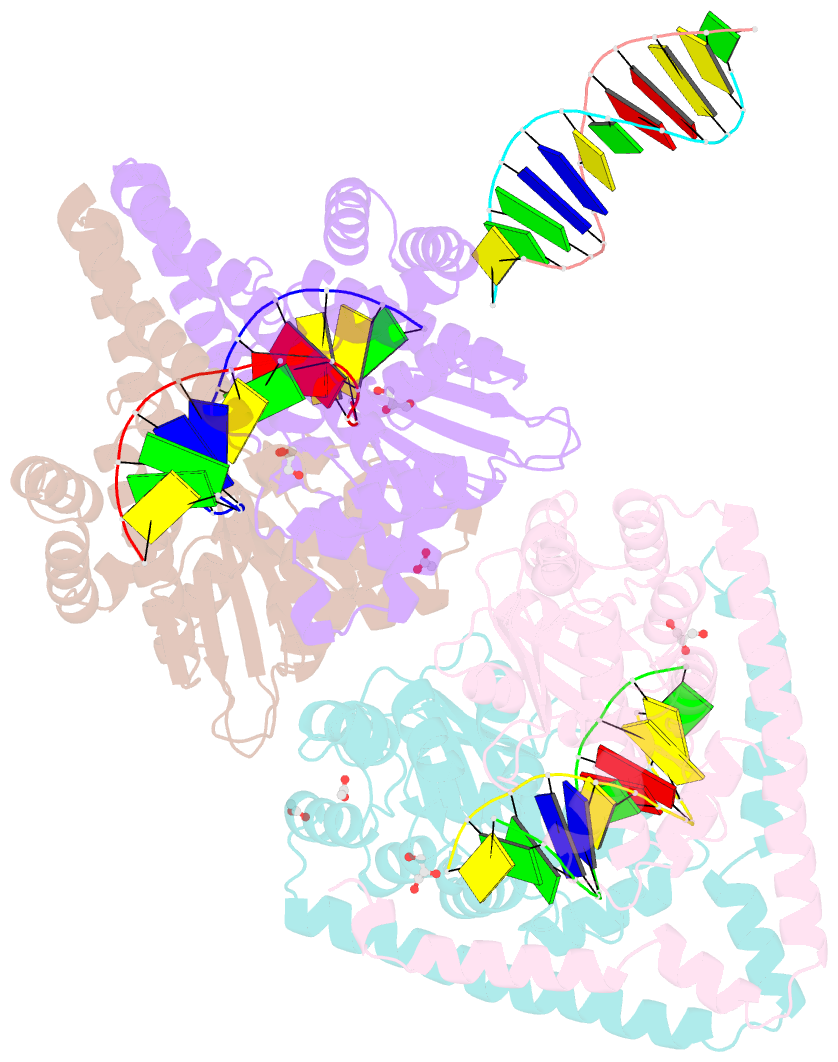
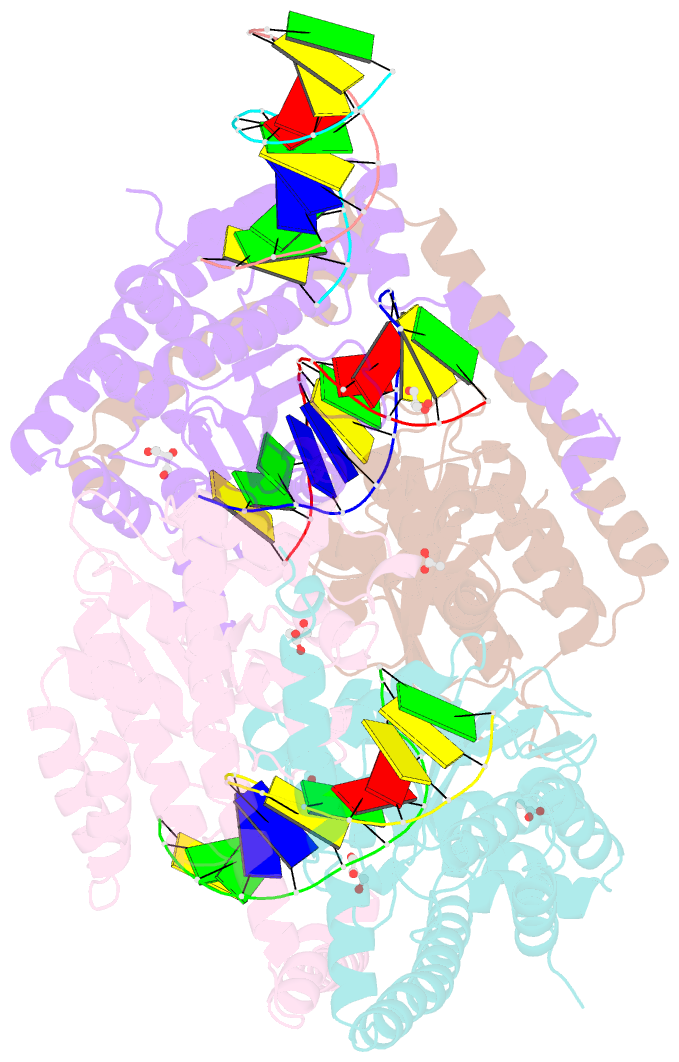
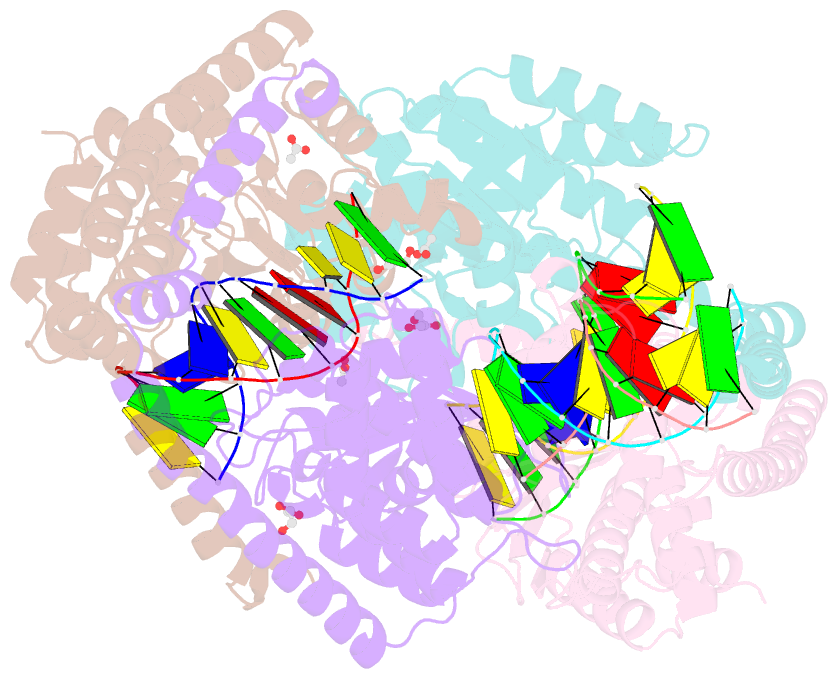
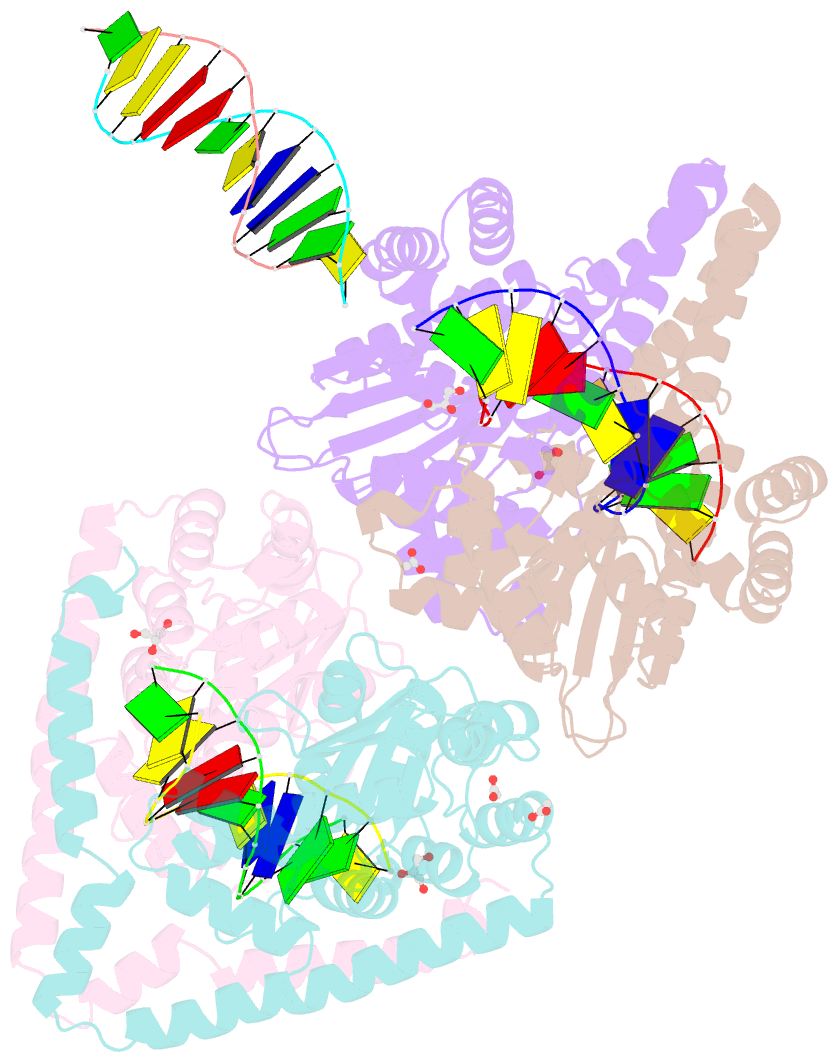
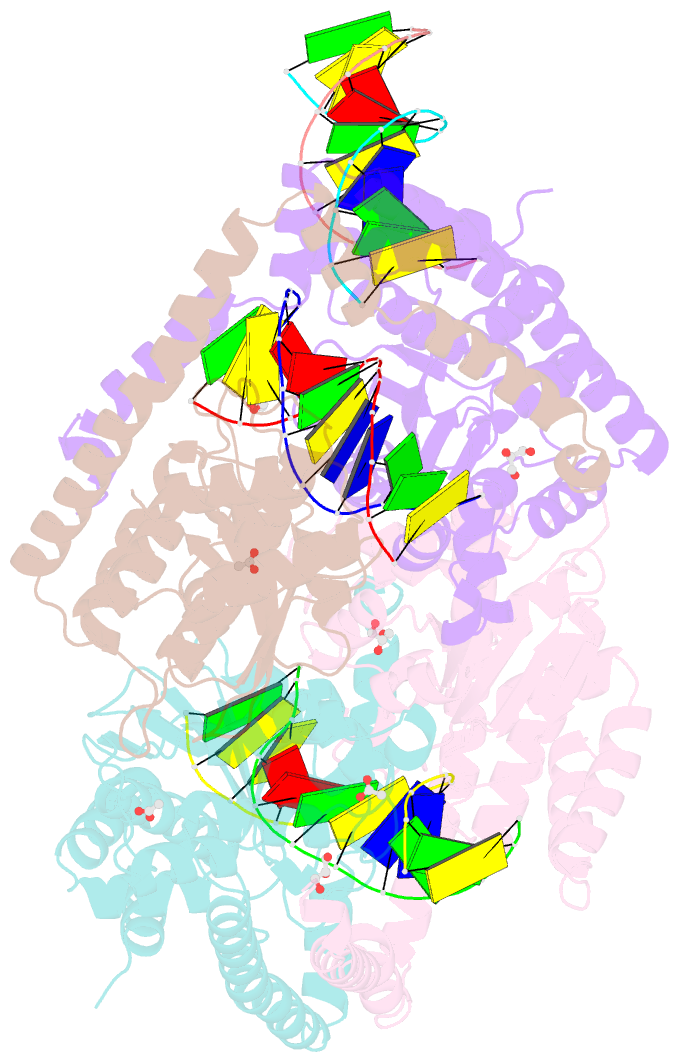
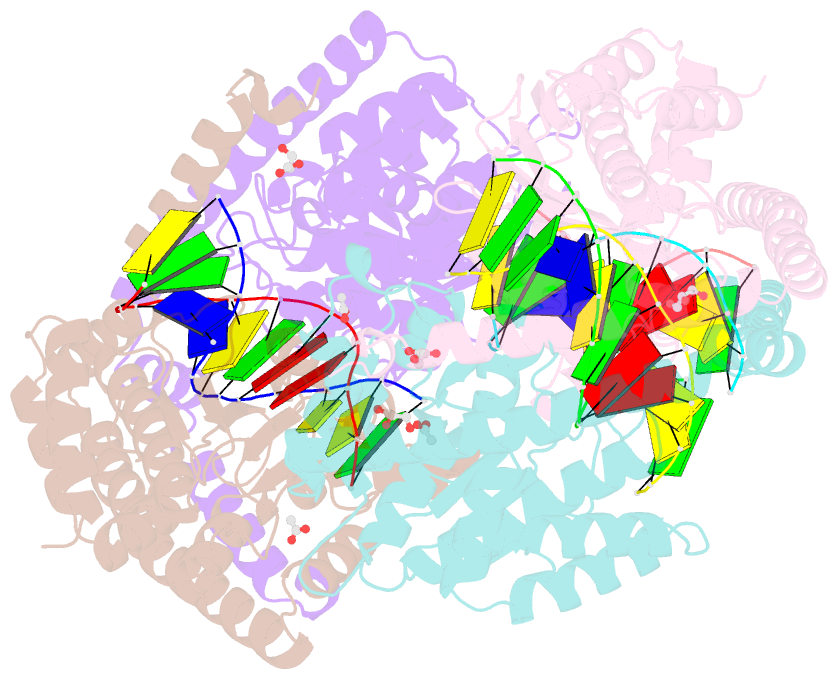
- Crystal structural analysis of hindiii restriction endonuclease in complex with cognate DNA at 2.0 angstrom resolution
- Reference
- Watanabe N, Takasaki Y, Sato C, Ando S, Tanaka I (2009): "Structures of restriction endonuclease HindIII in complex with its cognate DNA and divalent cations." Acta Crystallogr.,Sect.D, 65, 1326-1333. doi: 10.1107/S0907444909041134.
- Abstract
- The three-dimensional crystal structures of HindIII bound to its cognate DNA with and without divalent cations were solved at 2.17 and 2.00 A resolution, respectively. HindIII forms a dimer. The structures showed that HindIII belongs to the EcoRI-like (alpha-class) subfamily of type II restriction endonucleases. The cognate DNA-complex structures revealed the specific DNA-recognition mechanism of HindIII by which it recognizes the palindromic sequence A/AGCTT. In the Mg(2+) ion-soaked structure the DNA was cleaved and two ions were bound at each active site, corresponding to the two-metal-ion mechanism.