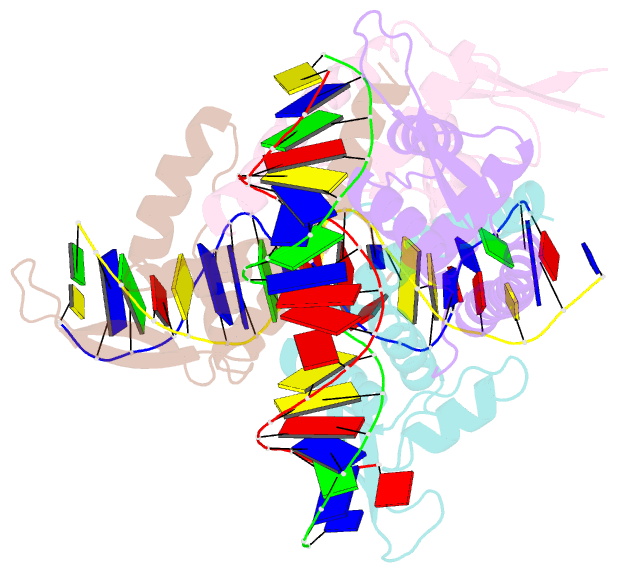
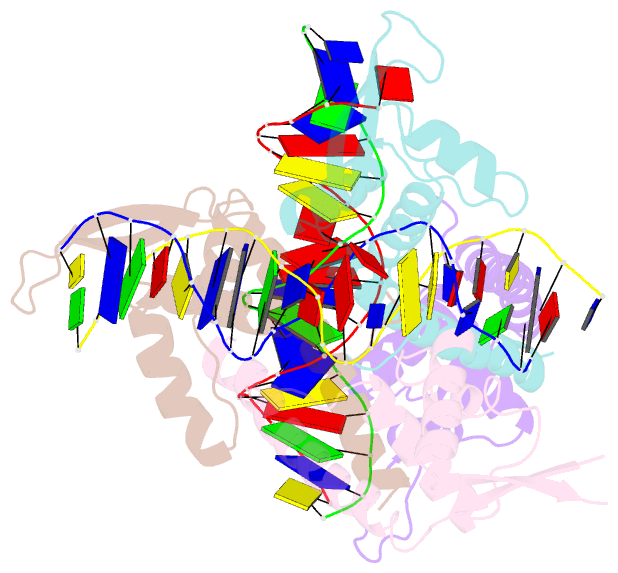
Summary information and primary citation
- PDB-id
- 2efw; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- replication-DNA
- Method
- X-ray (2.5 Å)
- Summary
- Crystal structure of the rtp:nrb complex from bacillus subtilis
- Reference
- Vivian JP, Porter CJ, Wilce JA, Wilce MCJ (2007): "An asymmetric structure of the Bacillus subtilis replication terminator protein in complex with DNA." J.Mol.Biol., 370, 481-491. doi: 10.1016/j.jmb.2007.02.067.
- Abstract
- In Bacillus subtilis, the termination of DNA replication via polar fork arrest is effected by a specific protein:DNA complex formed between the replication terminator protein (RTP) and DNA terminator sites. We report the crystal structure of a replication terminator protein homologue (RTP.C110S) of B. subtilis in complex with the high affinity component of one of its cognate DNA termination sites, known as the TerI B-site, refined at 2.5 A resolution. The 21 bp RTP:DNA complex displays marked structural asymmetry in both the homodimeric protein and the DNA. This is in contrast to the previously reported complex formed with a symmetrical TerI B-site homologue. The induced asymmetry is consistent with the complex's solution properties as determined using NMR spectroscopy. Concomitant with this asymmetry is variation in the protein:DNA binding pattern for each of the subunits of the RTP homodimer. It is proposed that the asymmetric "wing" positions, as well as other asymmetrical features of the RTP:DNA complex, are critical for the cooperative binding that underlies the mechanism of polar fork arrest at the complete terminator site.