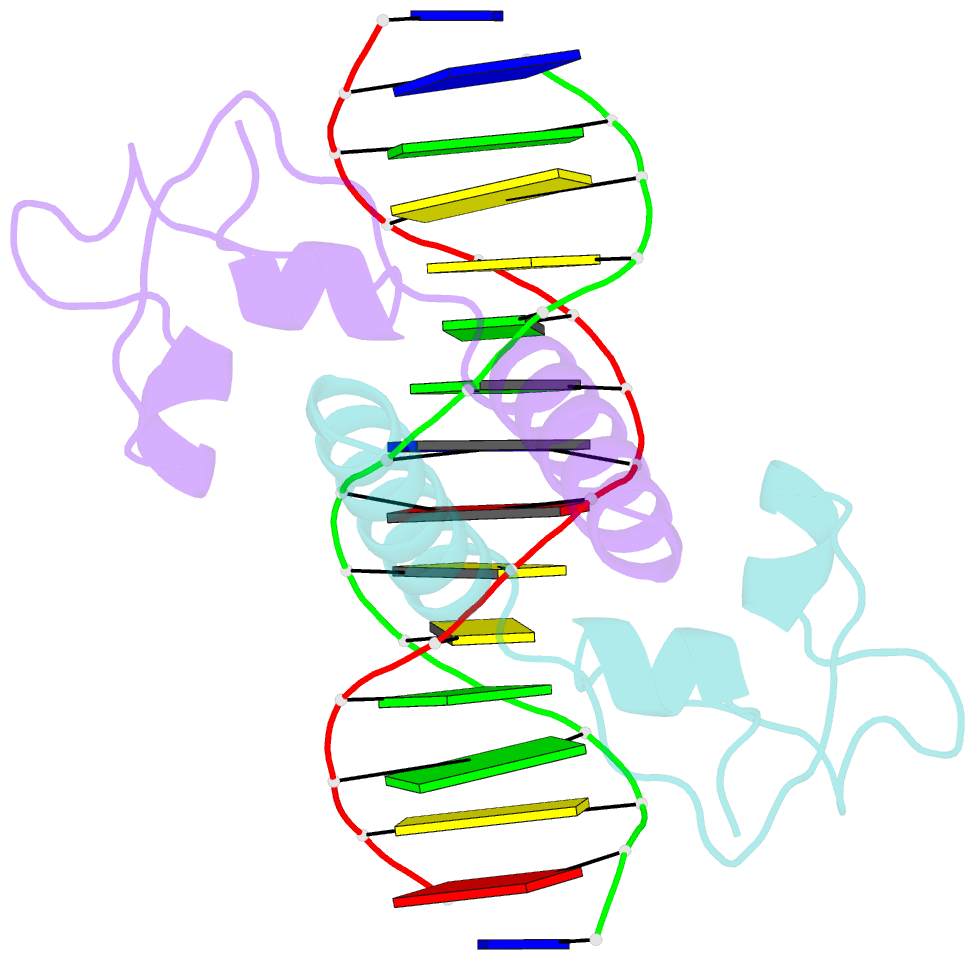
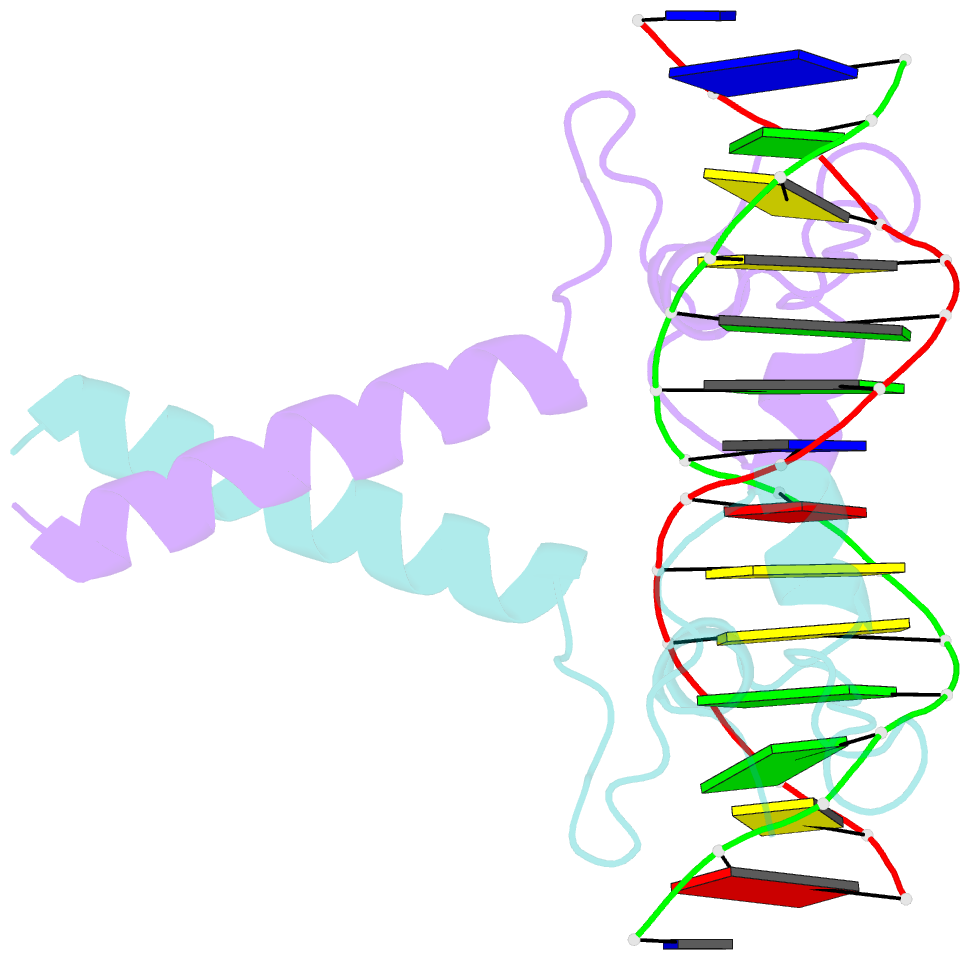
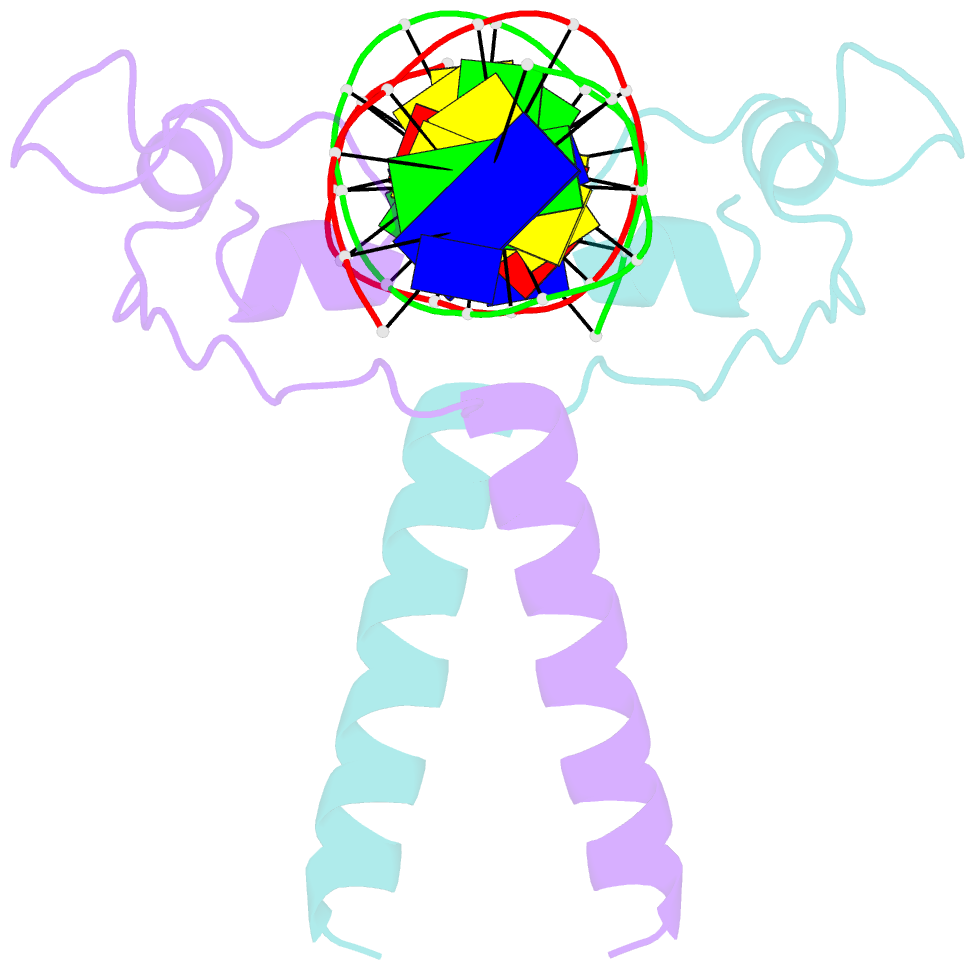
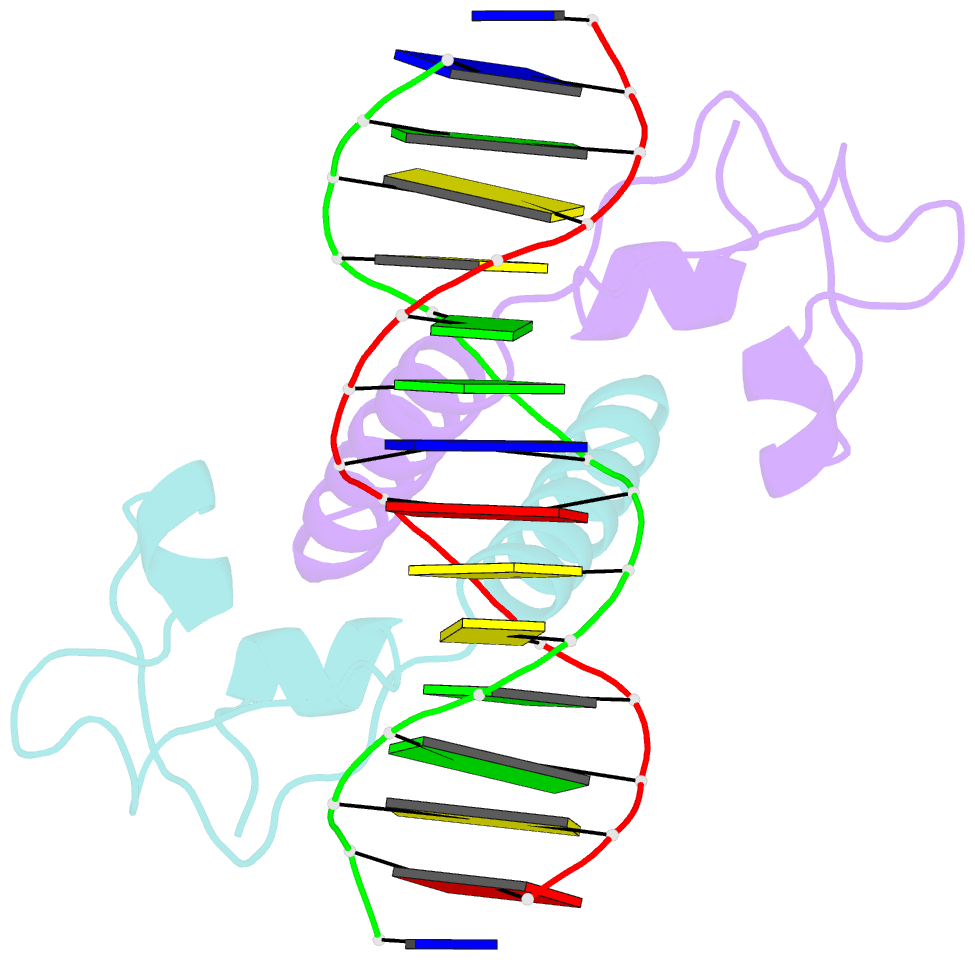
Summary information and primary citation
- PDB-id
- 2erg; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transcription activator-DNA
- Method
- X-ray (3.15 Å)
- Summary
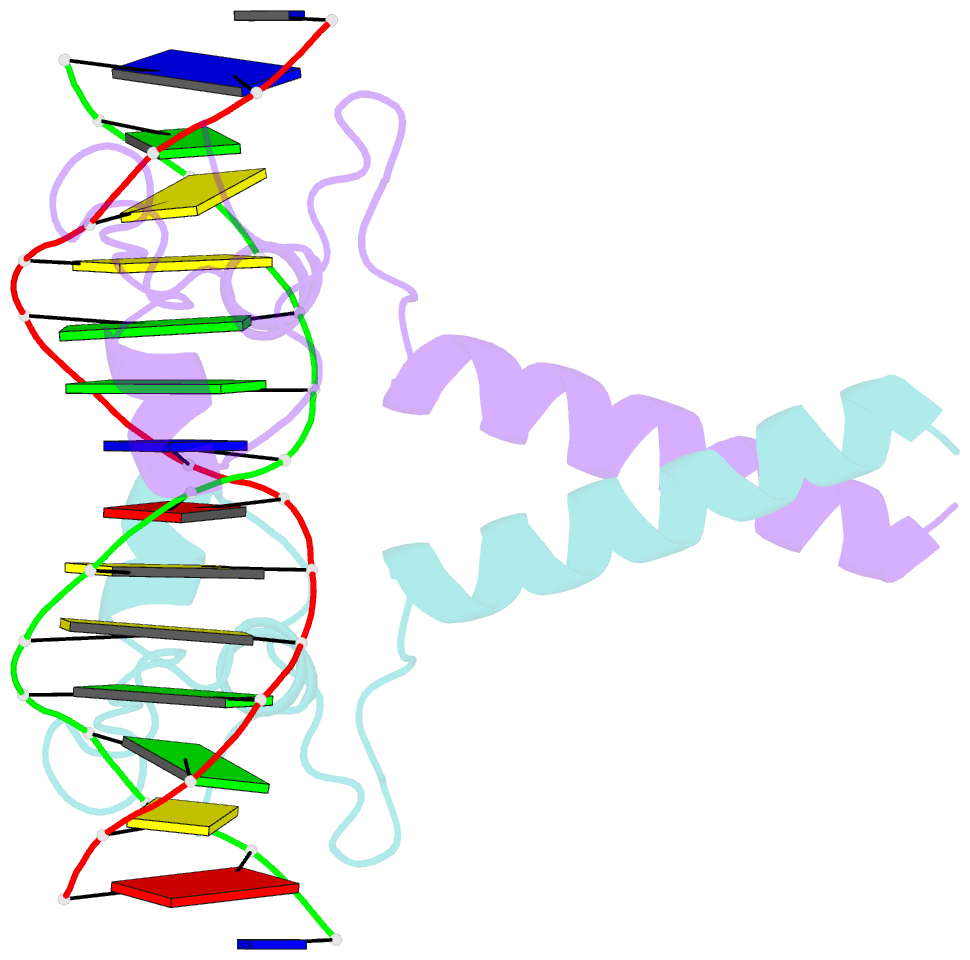
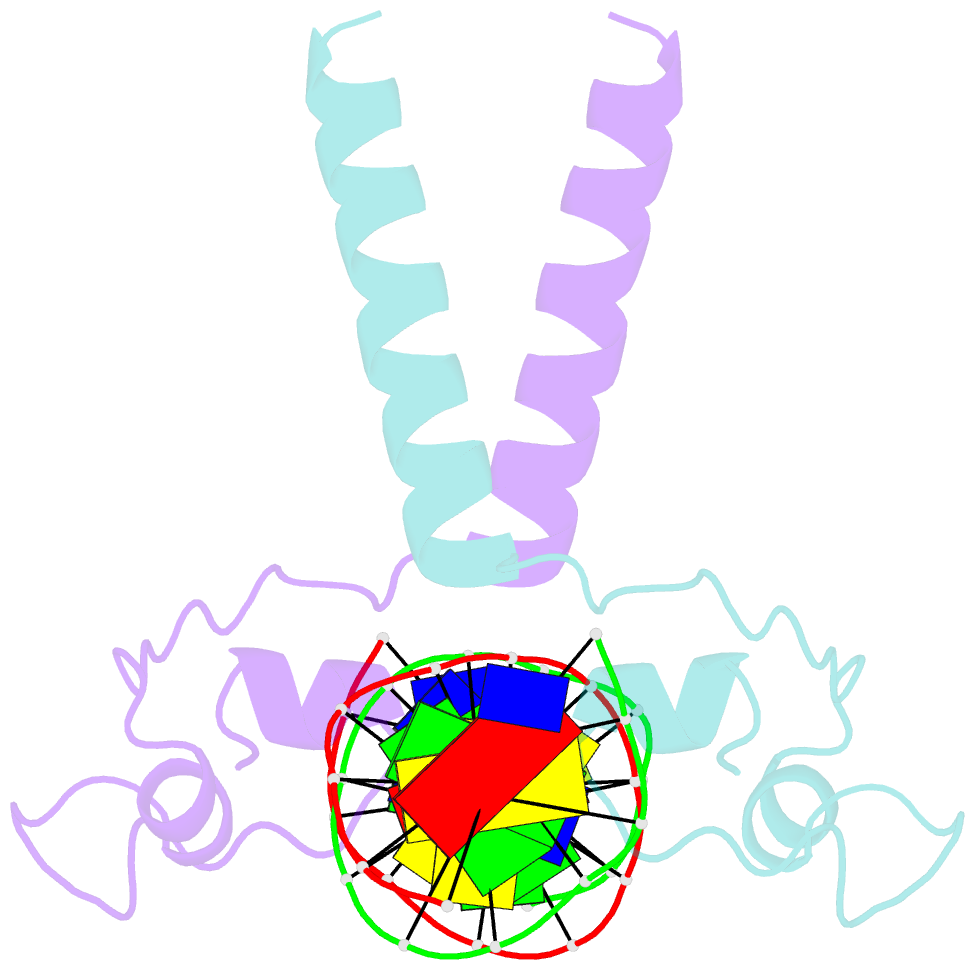
- Crystal structure of leu3 DNA-binding domain with a single h50c mutation complexed with a 15mer DNA duplex
- Reference
- Fitzgerald MX, Rojas JR, Kim JM, Kohlhaw GB, Marmorstein R (2006): "Structure of a Leu3-DNA complex: recognition of everted CGG half-sites by a Zn2Cys6 binuclear cluster protein." Structure, 14, 725-735. doi: 10.1016/j.str.2005.11.025.
- Abstract
- Gal4 is the prototypical Zn2Cys6 binuclear cluster transcriptional regulator that binds as a homodimer to DNA containing inverted CGG half-sites. Leu3, a member of this protein family, binds to everted (opposite polarity to inverted) CGG half-sites, and an H50C mutation within the Leu3 Zn2Cys6 binuclear motif abolishes its transcriptional repression function without impairing DNA binding. We report the X-ray crystal structures of DNA complexes with Leu3 and Leu3(H50C) and solution DNA binding studies of selected Leu3 mutant proteins. These studies reveal the molecular details of everted CGG half-site recognition, and suggest a role for the H50C mutation in transcriptional repression. Comparison with the Gal4-DNA complex shows an unexpected conservation in the DNA recognition mode of inverted and everted CGG half-sites, and points to a critical function of a linker region between the Zn2Cys6 binuclear cluster and dimerization regions in DNA binding specificity. Broader implications of these findings are discussed.